- EN - English
- CN - 中文
A Protocol to Depict the Proteolytic Processes Using a Combination of Metal–Organic Materials (MOMs), Electron Paramagnetic Resonance (EPR), and Mass Spectrometry (MS)
组合使用金属有机材料 (MOM)、电子顺磁共振 (EPR) 和质谱 (MS) 来描述蛋白水解过程的方案
发布: 2024年01月05日第14卷第1期 DOI: 10.21769/BioProtoc.4909 浏览次数: 1731
评审: Thomas SchmidtNeha Nandwani

相关实验方案
利用靶向GFP或mCherry的纳米抗体在出芽酵母中一步生成AlissAID条件性敲降菌株
Yoshitaka Ogawa [...] Takumi Kamura
2024年06月20日 2009 阅读
Abstract
Proteolysis is a critical biochemical process yet a challenging field to study experimentally due to the self-degradation of a protease and the complex, dynamic degradation steps of a substrate. Mass spectrometry (MS) is the traditional way for proteolytic studies, yet it is challenging when time-resolved, step-by-step details of the degradation process are needed. We recently found a way to resolve the cleavage site, preference/selectivity of cleavage regions, and proteolytic kinetics by combining site-directed spin labeling (SDSL) of protein substrate, time-resolved two-dimensional (2D) electron paramagnetic resonance (EPR) spectroscopy, protease immobilization via metal–organic materials (MOMs), and MS. The method has been demonstrated on a model substrate and protease, yet there is a lack of details on the practical operations to carry out our strategy. Thus, this protocol summarizes the key steps and considerations when carrying out the EPR/MS study on proteolytic processes, which can be generalized to study other protein/polypeptide substrates in proteolysis. Details for the experimental operation and cautions of each step are reported with figures illustrating the concepts. This protocol provides an effective approach to understanding the proteolytic process with the advantages of offering time-resolved, residue-level resolution of structural basis underlying the process. Such information is important for revealing the cleavage site and proteolytic mechanisms of unknown proteases. The advantage of EPR, probing the target substrate regardless of the complexities caused by the proteases and their self-degradation, offers a practically effective, rapid, and easy-to-operate approach to studying proteolysis.
Key features
• Combining protease immobilization, EPR, spin labeling, and MS experimental methods allows for the analysis of proteolysis process in real time.
• Reveals cleavage site, kinetics of product generation, and preference of cleavage regions via time-resolved SDSL-EPR.
• MS confirms EPR findings and helps depict the sequences and populations of the cleaved segments in real time.
• The demonstrated method can be generalized to other proteins or polypeptide substrates upon proteolysis by other proteases.
Graphical overview
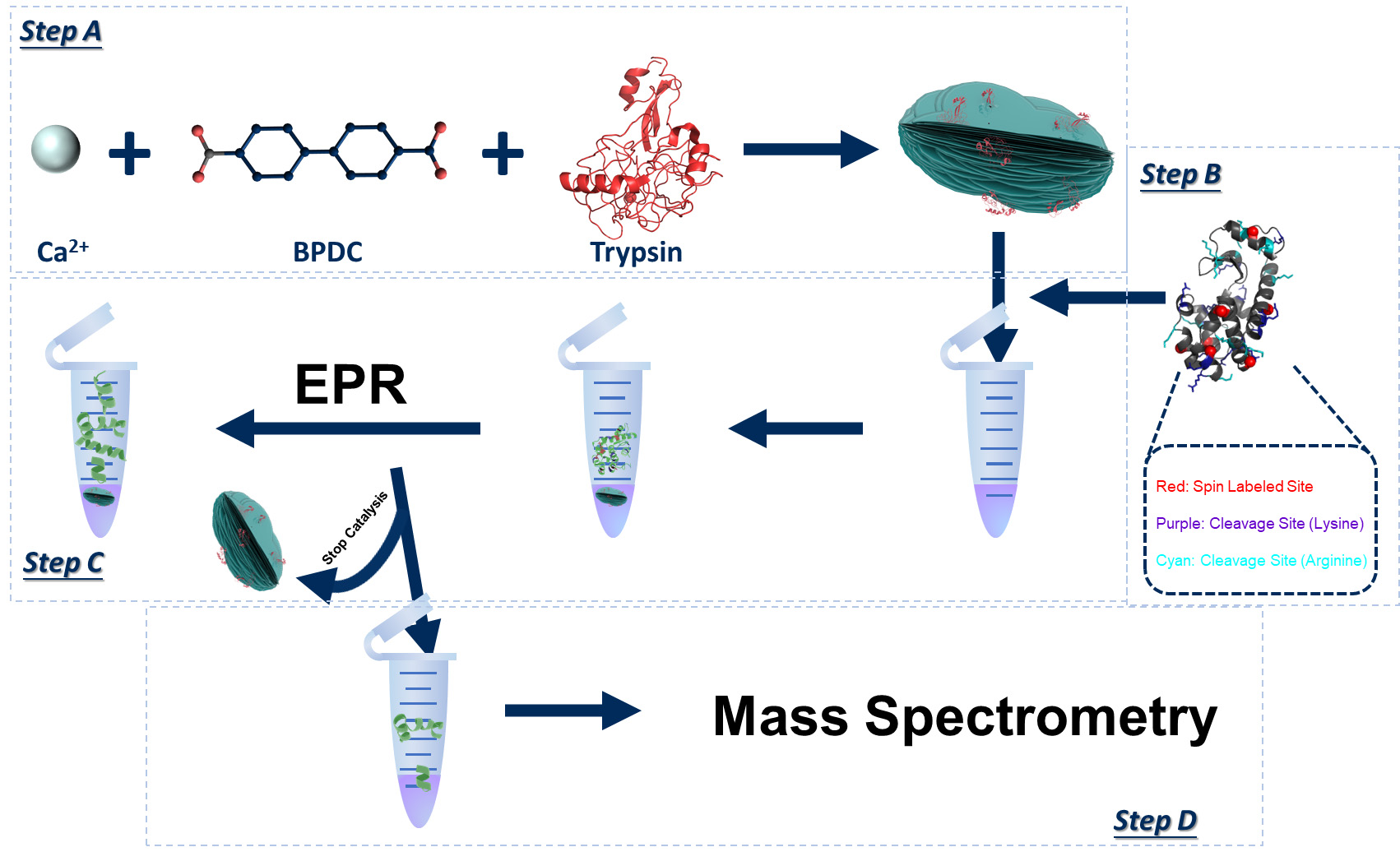
Background
Proteolysis is a critical cellular/biochemical process receiving extensive research attention that has found wide applications in industry and biomedicine [1–6]. Most current progress has been focused on the structures [7–9], cleavage sites [10–13], and proteolytic mechanisms of commonly seen proteases [14–17]. Meanwhile, a knowledge gap still remains regarding the molecular level details of the step-by-step, proteolytic actions on the substrate polypeptides/proteins. Bridging this gap is challenging because it requires revealing, in real time, the details of the reaction mixture, a complex ensemble containing peptide pieces with constantly changing lengths and populations. Perhaps the most feasible practice to probe the lengths and populations of the broken peptide sequences of the entire ensemble would be tandem mass spectrometry (MS/MS or MS2) [18–21], which also resolves amino acid sequence [22–24]. However, MS does not conveniently provide time-resolved information due to the change in product size and population as well as the self-degradation of the protease, which may create additional peptide sequences in the reaction mixture in real time, complicating the mass analysis and even the proteolytic kinetics due to the loss in protease over time. Thus, MS is often applied to analyze the final products after a certain stage (or upon completion) of a proteolytic reaction.
Recently, we found it possible to track the proteolytic process using site-directed spin labeling (SDSL) in combination with electron paramagnetic resonance (EPR) spectroscopy, which is sensitive only to the spin-labeled peptide truncations regardless of the presence of the protease and other broken peptide pieces [25]. This is because EPR line width is sensitive to the molecular size of the labeled polypeptides, which can be tracked via time-resolved, two-dimensional (2D) EPR [26–27]. We also found that immobilizing proteases on the surface of metal–organic materials (MOMs) crystals via co-crystallization allowed for the separation of protease from the soluble reaction mixture during a proteolytic reaction, which contains intact and broken peptides, by centrifugation [28–30]. The soluble portion can then be subjected to MS/MS study to determine the peptide sequences of the entire ensemble. Thus, we proposed a combination of MOMs, EPR, and MS to reveal the global (via MS) and local (via EPR) structural information of the peptide substrates in real time upon proteolysis, which ended up with a dynamic movie depicting the cleavage process [25]. As that was the first time that such a combination was applied to study proteases, we believe it is necessary to summarize the practical protocols to carry out such a complicated study. This protocol is based on our recent publication and utilized the data therein to demonstrate the protocol [25].
Materials and reagents
Reagents
Biphenyl-4,4′-dicarboxylic acid (BPDC-COOH) (Sigma-Aldrich, catalog number: 225266)
NaOH (Sigma-Aldrich, catalog number: 221465)
2-Isopropanol (Fisher Scientific, catalog number: A416-4)
CaCl2 (Sigma-Aldrich, catalog number: C4901)
Trypsin (Sigma-Aldrich, catalog number: T1426)
1-Oxyl-2,2,5,5-tetramethyl-∆3-(methanesulfonyloxymethyl)pyrroline (MTSL) (Toronto Research Chemicals, catalog number: O872400)
Acetonitrile (Fisher Scientific, catalog number: A21-200)
3-(N-Morpholino)propanesulfonic acid, 4-Morpholinepropanesulfonic acid (MOPS) (Sigma-Aldrich, catalog number: M1254)
NaCl (Sigma-Aldrich, catalog number: S9888)
4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES) (Sigma-Aldrich, catalog number: H3375)
Solutions
Spin buffer (pH 6.8) (see Recipes)
HEPES buffer (pH 7.4) (see Recipes)
Recipes
Spin buffer (pH 6.8)
Reagent Final concentration Quantity MOPS 50 mM 10.46 g NaCl 25 mM 1.46 g H2O n/a 1 L Total n/a 1 L HEPES buffer (pH 7.4)
Reagent Final concentration Quantity HEPES 50 mM 11.92 g NaCl 50 mM 2.92 g H2O n/a 1 L Total n/a 1 L
Laboratory supplies
Beaker (100 mL) (Fisher Scientific, catalog number: 02-540N)
Magnetic spin bar (VWR, catalog number: 58948-080)
Buchner funnel (Sigma-Aldrich, catalog number: Z247340)
Side arm flask (Sigma-Aldrich, catalog number: Z740685)
Microcentrifuge tube (Fisher Scientific, catalog number: 02-682-004)
Amicon spin concentrator (Millipore Sigma, catalog number: ACS 501024)
Borosilicate capillary tube (DWK Life Sciences, catalog number: 13-707-47)
Filter paper (Millipore Sigma, catalog number: WHA1093125)
Equipment
Oven, sample drying (Global Industrial, catalog number: T9FB918772)
Stirring hot plate, mixing (Thermo Scientific, catalog number: SP88854100)
Vortex, mixing (VWR, catalog number: 10153-838)
Nutation mixer, incubation (VWR, catalog number: 82009-202)
Centrifuge, separation (Thermo Scientific, catalog number: 75887203)
EPR, dynamic monitoring (Bruker, model: ESC-106)
Mass spectrometer, peptide mass (Water, model: SYNAPT MS)
Ultra performance liquid chromatography (UPLC), peptide separation (ACQUITY UPLC I-class System)
Software and datasets
Software and algorithms
Multi-component for CW EPR spectral simulation (https://sites.google.com/site/altenbach/labview-programs/epr-programs/multicomponent?authuser=0)
Confirm peptide sequence ProteinLynx Global SERVER 3.0.3
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Li, Q., Lenertz, M., Armstrong, Z., MacRae, A., Feng, L., Ugrinov, A. and Yang, Z. (2024). A Protocol to Depict the Proteolytic Processes Using a Combination of Metal–Organic Materials (MOMs), Electron Paramagnetic Resonance (EPR), and Mass Spectrometry (MS). Bio-protocol 14(1): e4909. DOI: 10.21769/BioProtoc.4909.
分类
生物物理学 > EPR波谱
生物化学 > 蛋白质 > 降解
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link