- EN - English
- CN - 中文
Workflow for High-throughput Screening of Enzyme Mutant Libraries Using Matrix-assisted Laser Desorption/Ionization Mass Spectrometry Analysis of Escherichia coli Colonies
使用基质辅助激光解吸/电离质谱分析大肠杆菌菌落高通量筛选酶突变体文库的工作流程
发布: 2023年11月05日第13卷第21期 DOI: 10.21769/BioProtoc.4862 浏览次数: 2224
评审: Neha NandwaniRama Reddy GoluguriAmit Kumar Dey
Abstract
High-throughput molecular screening of microbial colonies and DNA libraries are critical procedures that enable applications such as directed evolution, functional genomics, microbial identification, and creation of engineered microbial strains to produce high-value molecules. A promising chemical screening approach is the measurement of products directly from microbial colonies via optically guided matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). Measuring the compounds from microbial colonies bypasses liquid culture with a screen that takes approximately 5 s per sample. We describe a protocol combining a dedicated informatics pipeline and sample preparation method that can prepare up to 3,000 colonies in under 3 h. The screening protocol starts from colonies grown on Petri dishes and then transferred onto MALDI plates via imprinting. The target plate with the colonies is imaged by a flatbed scanner and the colonies are located via custom software. The target plate is coated with MALDI matrix, MALDI-MS analyzes the colony locations, and data analysis enables the determination of colonies with the desired biochemical properties. This workflow screens thousands of colonies per day without requiring additional automation. The wide chemical coverage and the high sensitivity of MALDI-MS enable diverse screening projects such as modifying enzymes and functional genomics surveys of gene activation/inhibition libraries.
Key features
• Mass spectrometry analyzes a range of compounds from E. coli colonies as a proxy for liquid culture testing enzyme mutant libraries.
• Colonies are transferred to a MALDI target plate by a simple imprinting method.
• The screen compares the ratio among several products or searches for the qualitative presence of specific compounds.
• The protocol requires a MALDI mass spectrometer.
Graphical overview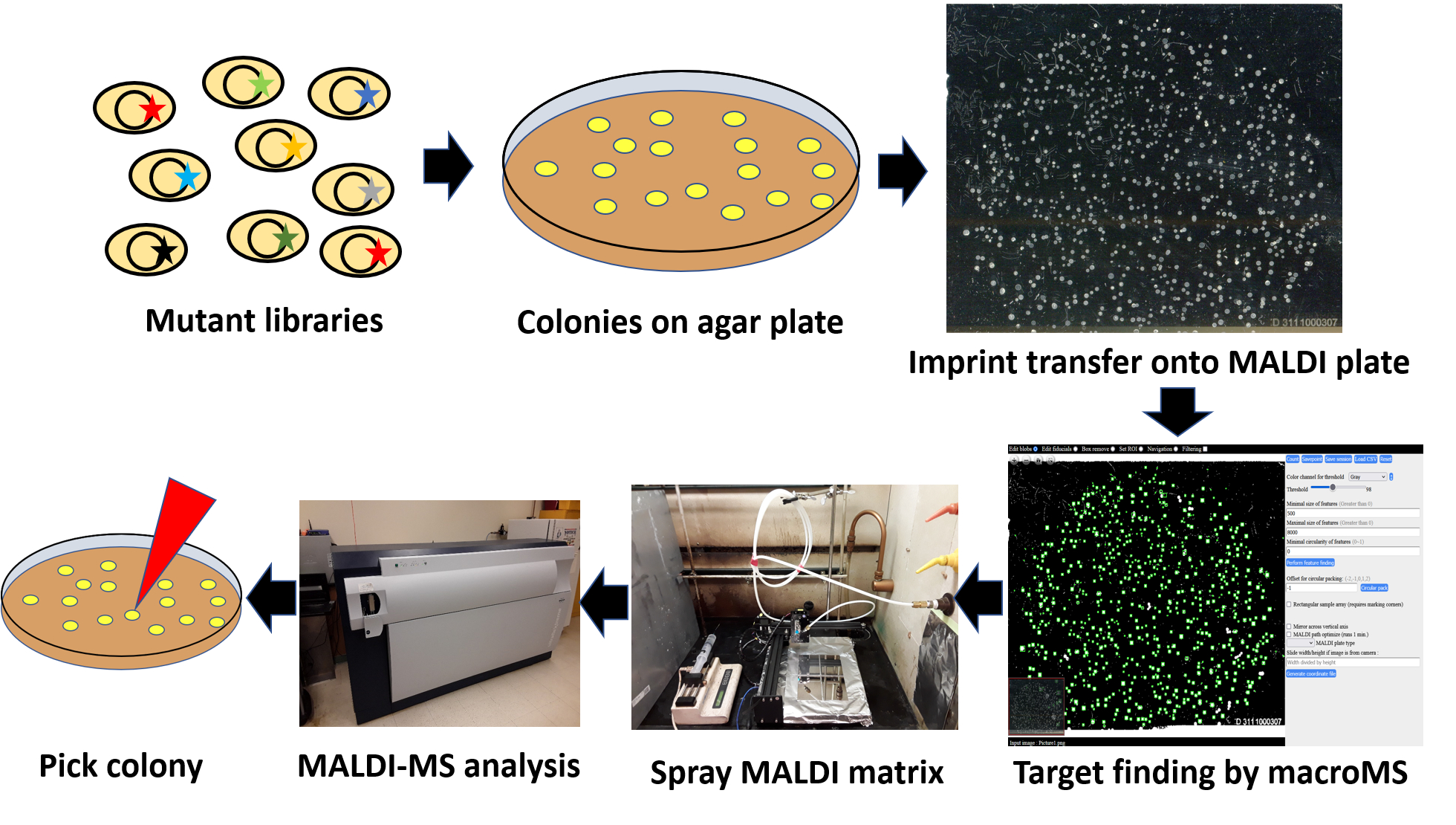
Overview of the MALDI-MS analysis of microbial colonies for screening mutant libraries. Microbial cells containing a mutant library for enzymes/metabolic pathways are first grown in agar. The colonies are then imprinted onto a MALDI target plate using a filter paper intermediate. An optical image of the MALDI target plate is analyzed by custom software to find the locations of individual colonies and direct subsequent MALDI-MS analyses to the selected colonies. After applying MALDI matrix onto the target plate, MALDI-MS analysis of the colonies is performed. Colonies showing the desired product profiles are found by data analysis via the software, and the colonies are picked for downstream analysis.
Background
Directed evolution is an iterative process of mutagenesis and phenotype screening to achieve incremental improvements towards desired characteristics (Arnold, 2018). The method is used widely for creating efficient biocatalysts producing drugs, fuels, food, and industrial products (Turner, 2009; Dietrich et al., 2010; Wang et al., 2021). Lack of high-throughput methods for phenotype screening can be a bottleneck for directed evolution (Zeymer and Hilvert, 2018). Chromogenic conversion, pH change, and fluorescence readings can be used for phenotype screening; however, they are often indirect measurements of the desired phenotype (Körfer et al., 2018; Alfaro-Chávez et al., 2019; Minges et al., 2020). A promising approach is mass spectrometry (MS), which provides label-free measurement of a wide variety of analytes. Acoustic ejection mass spectrometry is the state-of-the-art high-throughput screening technology that applies an acoustic pulse to transfer liquid samples to an MS device, achieving a subsecond analytical cycle (Liu et al., 2020; Zhang et al., 2021). However, it requires extensive automation systems for sample creation such as biochemical reactions, cell culture, liquid transfer, and storage (Simon et al., 2021). Such robotic systems are not commonly available to many labs. Alternatively, matrix-assisted laser desorption/ionization MS (MALDI-MS) is a widely available MS system that boasts high throughput, relatively simple sample preparation, high salt tolerance, and wide chemical coverage (Fournier et al., 2008; Grove et al., 2011; Singhal et al., 2015). Previously, the optically guided MALDI-MS method was applied to perform high-throughput screening of microbial colonies for engineering multistep enzyme reactions (Si et al., 2017). In this workflow, hundreds of agar colonies are transferred to a MALDI plate by imprinting, and MALDI-MS of the scattered colonies is performed using the coordinates reported by microMS image analysis software. This work successfully enables individual colonies to serve as individual reaction vessels for phenotyping, which is a promising alternative to the complex automation system needed for performing and processing large numbers of liquid cultures. For routine analysis of a larger number of microbial colonies, the current protocol simplifies steps for imaging, instrument setup, software operation, and downstream data analysis.
The simplification of the workflow is enabled by macroMS, a newly created software package that is freely available. The macroMS is the adaptation of a single-cell microMS package and it was created for high-throughput screening of macroscopic samples larger than 200 μm (Comi et al., 2017; Choe et al., 2021). The macroMS package enables rapid imaging by flatbed scanner and camera, user-friendly tools for image analysis and optical correction, simple MS instrument training, and data analysis/visualization for isolating colonies of interest. Based on macroMS, this protocol describes a simplified end-to-end colony screening workflow by MALDI-MS, including growing and preparing colonies for the MALDI-MS screen, using the macroMS software to set up and perform MALDI-MS analysis, performing data analysis, and picking colonies of interest. This workflow can be used to efficiently screen mutant libraries for modifying a wide variety of enzymes or metabolic pathways that form compounds on colonies that are detectable by MALDI-MS. While the workflow has been developed using the Bruker ultraflextreme mass spectrometer, it can be adapted to other instruments, although this would require adapting several of the software and instrument operation steps. The limitations of the workflow are: 1) it works with nonvolatile compounds that do not evaporate in the vacuum environment of MALDI-MS, 2) it is a semiquantitative MALDI-MS screen that uses the ratio between observed MALDI-MS peaks, and 3) the macroMS data analysis tool limits the width of the mass range of the mass spectrum to be under m/z 1,000.
As the equipment to spray matrix onto the slides is not commonly available in many laboratories, the end of the protocol includes the construction of a spraying device for applying MALDI matrix onto four extra-large MALDI target plates (i.e., 105 mm × 75 mm). MALDI matrix application is a commonly performed step for MALDI imaging, where airbrush, sublimation chamber, and commercial automatic sprayer systems have been used to coat samples with the compounds (Hankin et al., 2007; Ye et al., 2013; Andersen et al., 2020). However, few devices have the capacity for processing wide target plates holding the large number of colonies needed for high-throughput screening. Also, while MALDI-MS has become commonly available in universities and industrial settings, facilities often lack the capability and experience with MALDI imaging and therefore do not have devices for MALDI matrix coating. If other alternatives are available, they can be used; otherwise, the system described here provides the capability to simultaneously prepare four large MALDI plates as needed for high-throughput screens and reducing variability.
The workflow was successfully used to screen mutant libraries for an acyl-ACP thioesterase and an acyl-ACP desaturase. Acyl-ACP thioesterase cleaves the thioester bond in acyl-ACPs in the type II fatty acid synthesis pathway, deciding the chain length of the resulting free fatty acids. Acyl-ACP thioesterase from the California bay laurel (UcFatB2) expressed in E. coli produces dodecanoic acid as a primary product and tetradecanoic acid as a secondary product. The workflow compared the ratio between dodecanoic acid and tetradecanoic acid measured from colonies that express UcFatB2 random mutant library and found variants that have higher ratio for dodecanoic acid (Jindra et al., 2023). Acyl-ACP desaturase removes two hydrogens from a single bond between two carbons (C — C) resulting in double bond (C = C) in fatty acyl-ACP, deciding the location of the double bond in unsaturated fatty acid. Acyl-ACP desaturase from the Black-eyed Susan vine (TaDes) expressed in E. coli produces C16:1 Δ6 as a primary product and C16:1 Δ8 acid as a secondary product. By combining with a simple ozone treatment step, the workflow compared the ratio between the ozonolysis products of C16:1 Δ6 and C16:1 Δ8 measured from colonies that express TaDes random mutant library. The screen found variants that produce C16:1 Δ8 as a primary product (Choe et al., 2023). The protocol covers the workflow demonstrated using the thioesterase screen.
Materials and reagents
Prespotted AnchorChip adapter (Bruker Daltonics, catalog number: 221598), four custom built 1.2 mm × 105 mm × 75 mm target plates, and copper conducting tape (3M, catalog number: 05012A-AB) (Note 1)
MTP target frame (Bruker Daltonics, catalog number: 8074115) and MTP 384 target plate polished steel (Bruker Daltonics, catalog number: 8280781)
Vacuum desiccator (SP Bel-Art, catalog number: F42020-0000)
5′ of 1/2″ tubing (McMaster-Carr, catalog number: 5648K33)
N-Phenyl-2-naphthylamine (PNA) (Sigma-Aldrich, catalog number: 178055)
7.25″ × 5″ × 1.875″ HDX hydrophilic sponge (QEP, catalog number: 70005-24)
6″ × 6″ × 3/8″ metal plate (McMaster-Carr, catalog number: 8983K212)
6″ × 6″ × 0.12″ metal plate (McMaster-Carr, catalog number: 8983K118)
A4 papers
Standard lab tape
Lighter
Push pins
Duct tape
Sharpie pen
Power strip
Isopropyl alcohol
Methanol
Glycerol
Delicate task wipes (Kimberly-Clark Professional, catalog number: 34155)
10% bleach
150 mm × 15 mm bacteriological Petri dish (Corning, catalog number: 351058)
Syringe filter, 0.2 μm Nylon (Thermo Scientific, catalog number: 726-2520)
90 mm filter unit, 500 mL (Thermo Scientific, catalog number: 569-0020)
Sterile 10 mL syringe, single use (BD, catalog number: 302995)
Sterile 1.5, 15, and 50 mL tubes and corresponding tube racks
Sterile 2, 20, and 1,000 μL pipette tips and corresponding adjustable volume pipettes
Sterile 5 and 50 mL serological pipettes and a pipetaid
Carbenicillin (Gold Biotechnology, catalog number: C-103-5)
Autoclaved 90 mm membrane filter, 0.22 μm (Millipore, catalog number: GVWP09050)
14 mL cell culture tubes (Greiner Bio-One, catalog number: 187262)
Sterile 250 and 1,000 mL autoclavable flasks (Pyrex, catalog number: 4980)
L-shaped cell spreader (VWR, catalog number: 76207-748)
Inoculating turntable
Ice bucket
4″ × 125′ Parafilm-M film (Bemis Company Inc, catalog number: PM996)
Gallon storage double zip bags, 26.8 cm × 27.3 cm (Walmart)
Plastic box for transporting MALDI target adapter
Aluminum foil
Tweezer
Spectrophotometer cuvette
10 nm gold colloid solution (BBI Solutions, catalog number: EM.GC10/4)
LB powder (Fisher Scientific, catalog number: BP9722-2)
Agar (BD, catalog number: 214030)
Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: IPTG-RO)
Glucose (Fisher Scientific, catalog number: D1610)
Autoclave tapes
Clock
Paper towel
Miniprep kit (QIAGEN, catalog number: 27104)
Chemical waste bottle
Cuffed sleeve lab coat
Disposable laboratory exam gloves
Biohazard trash can
Electrocompetent cells (i.e., RL08ara strain)
Mutant library plasmid (i.e., pTrc99a-*BTE)
Wild-type control plasmid (i.e., pTrc99a-WT BTE)
Acetonitrile (Thermo Fisher Scientific, catalog number: 51101) (Note 2)
Acetone (Fisher Scientific, catalog number: A929-1)
LB agar plates (see Recipes) (Note 3)
Liquid LB media (see Recipes)
PNA solution (per 20 mL) (see Recipes) (Note 2)
(Optional) materials for building MALDI matrix spray station
CNC machine (Huizhou Bachin Electronic Technology, catalog number: T-A4)
Windows PC compatible with the CNC machine
Chemical safety hood with working ports for nitrogen, air, vacuum, and electricity. The plastic barbed connector for the nitrogen port must be detachable by unscrewing, exposing the female opening for the pipe
Syringe pump (KDS Scientific, catalog number: KDS-100-CE)
Lab stand with a clamp
Two adjustable wrenches
Caliper
25′ of 1/4″ nylon tubing (McMaster-Carr, catalog number: 5548K75)
Two 50 mL syringes with Luer tapering (BD, catalog number: 309653)
1/4″ × 1/16″ stainless steel reducing union (Swagelok, catalog number: SS-400-6-1)
1/16″ OD × .020″ ID stainless steel tubing, 5 cm (IDEX, catalog number: U-101)
1/16″ OD × .030″ ID stainless steel tubing, 5 cm (IDEX, catalog number: U-115)
1/16″ Union Tee (Swagelok, catalog number: SS-100-3)
Two 1/16″ OD × .0155″ ID × 1.6″ NanoTightTM tube sleeves (IDEX, catalog number: F-242)
350 μm OD × 250 μm ID fused silica capillary (Polymicro, catalog number: 1068150026)
10-32 female-to-female Luer adapter (IDEX, catalog number: P-659)
1/16″ OD one-piece finger-tight fitting, 10-32 coned (IDEX, catalog number: F-120Y)
Gorilla glue (The Gorilla Glue Company)
(Optional) HTS MALDI adapter 1.0 mm (Bruker Daltonics, catalog number: 1847571)
(Optional) HTS MALDI plates (Bruker Daltonics, catalog number: 1833280)
(Optional) MTP target frame (Bruker Daltonics, catalog number: 8074115)
(Optional) MTP 384 target plate polished steel (Bruker Daltonics, catalog number: 8280781)
(Optional) 1/4″ tube OD × 3/8″ ID-18 NPTF male connector (Parker, catalog number: 68C-4-6)
(Optional) 1 mL glass syringe (Hamilton, catalog number: Gastight #1001)
Recipes
LB agar plates
Make 500 mL of 2× concentrated LB media (20 g of LB powder per 500 mL media).
Perform filter sterilization of the solution.
Add 250 mL of deionized water into a 1 L flask.
Add 7.5 g of agar into the flask.
Autoclave the flask for 20 min at 15 psi and 121 .
Immediately after completing the autoclave sterilization, transfer the flask to a biosafety hood.
Add 250 mL of the 2× LB media at room temperature into the flask, while swirling the flask.
Add carbenicillin (100 µg/mL), IPTG (0.1 mM), and glucose (1% w/v).
Thoroughly mix the media by swirling the flask.
Pour 40 mL of the media into each 150 mm Petri dish.
Leave the dishes completely open and dry for 30 min in the biosafety hood.
Put the dishes into plastic bags and store at 4 °C. During storage, the dishes should face upside down.
Liquid LB media (500 mL)
After dissolving 10 g of LB powder into deionized water (500 mL), perform filter sterilization of the solution using the vacuum filter unit.
PNA solution (per 20 mL)
Reagent Quantity PNA 100 mg Acetonitrile (Note 2) 20 mL
Equipment
PC with internet connectivity and Firefox or Chrome web browser
Flatbed scanner (EPSON, catalog number: Perfection V300 Photo)
Biosafety hood
Chemical safety hood
-20 °C freezer
-70 °C freezer
4 °C refrigerator
Autoclave
Cell culture incubator (Fisher Scientific, catalog number: 1600D)
Electroporator (Bio-Rad, catalog number: Gene Pulser Xcell)
Refrigerated centrifuge (Eppendorf, catalog number: 5810 R)
UV-Vis spectrophotometer (Thermo Scientific, catalog number: Genesys 10S)
Cell culture shaker (New Brunswick, catalog number: I 24) with clamps for 250 mL flasks
MALDI-ToF-MS (Bruker Daltonics, catalog number: ultrafleXtreme)
Ultrasonic bath sonicator (Cole Parmer, catalog number: 8891R)
Solid insert tray for sonicator bath (Branson, catalog number: 100410174)
Milli-Q water supply (ELGA LabWater, catalog number: PURELAB flex)
Ice supply
Electronic scale (Mettler Toledo, catalog number: PB4002-S)
(Optional) Acrylic nitrogen purge cabinets with nitrogen flow (Cleatech, catalog number: 1500-1-A)
(Optional) IR Thermometer (FLUKE, catalog number: 62MAX+) (Note 4)
Software
flexControl 3.4 (Build 135.12, Compass for flexSeries 1.4, Bruker Daltonics)
Bachin Draw (Huizhou Bachin Electronic Technology, http://www.bachinmaker.com)
Inkscape 1.0 (Inkscape, https://inkscape.org)
macroMS (Lab software, https://macroms.scs.illinois.edu/)
flexAnalysis 3 (Bruker Daltonics)
Control software for scanner
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Choe, K. and Sweedler, J. V. (2023). Workflow for High-throughput Screening of Enzyme Mutant Libraries Using Matrix-assisted Laser Desorption/Ionization Mass Spectrometry Analysis of Escherichia coli Colonies. Bio-protocol 13(21): e4862. DOI: 10.21769/BioProtoc.4862.
分类
系统生物学 > 基因组学 > 筛选
微生物学 > 异源表达系统 > 大肠杆菌
生物化学 > 蛋白质
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link