- EN - English
- CN - 中文
Isolation of Epithelial and Stromal Cells from Colon Tissues in Homeostasis and Under Inflammatory Conditions
在稳态和炎症条件下从结肠组织中分离上皮细胞和基质细胞
(*contributed equally to this work, § Technical contact) 发布: 2023年09月20日第13卷第18期 DOI: 10.21769/BioProtoc.4825 浏览次数: 5680
评审: Kazem NouriArundhati MehtaAnonymous reviewer(s)
Abstract
Inflammation of the gastrointestinal tract is a prevalent pathology in diseases such as inflammatory bowel disease (IBD). Currently, there are no therapies to prevent IBD, and available therapies to treat IBD are often sub-optimal. Thus, an unmet need exists to better understand the molecular mechanisms underlying intestinal tissue responses to damage and regeneration. The recent development of single-cell RNA (sc-RNA) sequencing-based techniques offers a unique opportunity to shed light on novel signaling pathways and cellular states that govern tissue adaptation or maladaptation across a broad spectrum of diseases. These approaches require the isolation of high-quality cells from tissues for downstream transcriptomic analyses. In the context of intestinal biology, there is a lack of protocols that ensure the isolation of epithelial and non-epithelial compartments simultaneously with high-quality yield. Here, we report two protocols for the isolation of epithelial and stromal cells from mouse and human colon tissues under inflammatory conditions. Specifically, we tested the feasibility of the protocols in a mouse model of dextran sodium sulfate (DSS)-induced colitis and in human biopsies from Crohn’s patients. We performed sc-RNA sequencing analysis and demonstrated that the protocol preserves most of the epithelial and stromal cell types found in the colon. Moreover, the protocol is suitable for immunofluorescence staining of surface markers for epithelial, stromal, and immune cell lineages for flow cytometry analyses. This optimized protocol will provide a new resource for scientists to study complex tissues such as the colon in the context of tissue damage and regeneration.
Key features
• This protocol allows the isolation of epithelial and stromal cells from colon tissues.
• The protocol has been optimized for tissues under inflammatory conditions with compromised cell viability.
• This protocol is suitable for experimental mouse models of colon inflammation and human biopsies.
Graphical overview
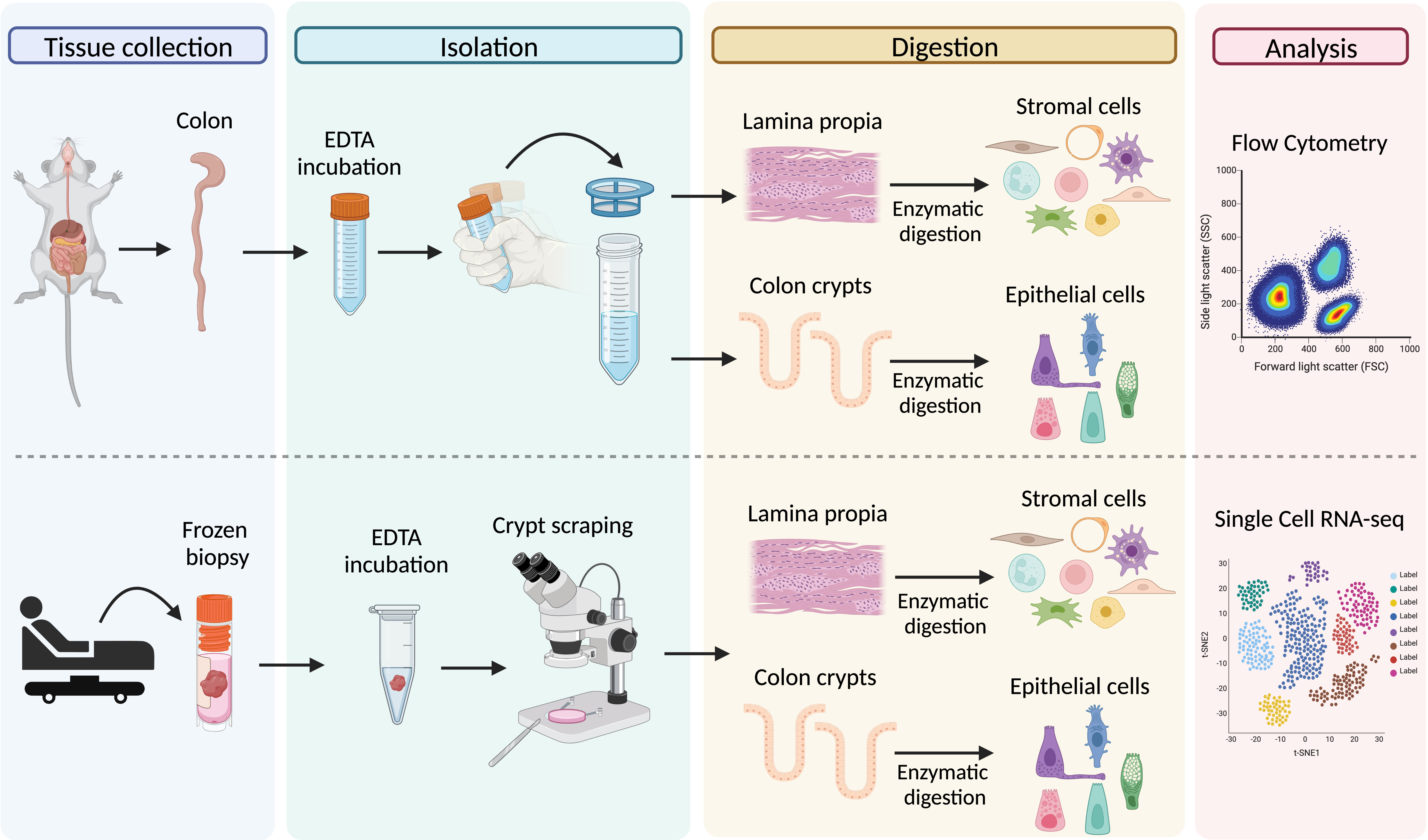
Graphical representation of the main steps for the processing of colon tissue from dextran sodium sulfate (DSS)-treated mice (upper panel) and frozen biopsies from Crohn’s patients (lower panel)
Background
Inflammatory bowel disease (IBD) is a chronic inflammatory condition of the gastrointestinal (GI) tract that encompasses two distinct pathologies: ulcerative colitis and Crohn’s disease. The global prevalence of IBD has been increasing since 2000 and now affects approximately 1.4 million individuals in America and 2.3 million in Europe (Ng et al., 2017). Current medical treatment for IBD involves the use of immunomodulatory agents to empirically dampen inflammation; however, up to one-third of patients fail initial treatment, and half of the initial responders will eventually relapse.
The interplay between the epithelial and stromal compartments supports homeostasis of the GI tract. Epithelial cells form a dynamic cellular layer with high regeneration potential that separates the luminal content from the underlying stromal tissue while supporting nutrient and water absorption. Simultaneously, the resident stromal cells of the lamina propria provide structural support, immunity against pathogens, and tolerance to the commensal microbiota. Breakdown of this equilibrium by dysregulation of host–microbiome interactions, defects in intestinal epithelial barrier permeability, and loss of immune tolerance can lead to the development of IBD (Maloy and Powrie, 2011). Therefore, it is important to consider the complex cellular heterogeneity when designing new experimental approaches to better understand the molecular mechanisms underlying disease.
The recent development of single-cell technologies has provided a finer picture of complex biology and deciphered heterogeneity present in tissue, enabling the identification of new cell populations, cellular interactions, and signaling pathways associated with pathology. However, these powerful approaches require the isolation of high-quality single cells from tissues to provide a representative and reliable tissue-associated cellular landscape. While the field of single-cell technology is moving fast, scientists have had to adapt tissue dissociation protocols depending on the cell types and contexts of interest. In the context of intestinal biology, the complex cellular heterogeneity of the gut mucosa and sensitivity to detachment-mediated cell death has hindered protocols for tissue dissociation that aim to represent the cellular repertoire of the original tissue. The main limitation is the fragility of the epithelial cells that cannot support most enzymes used to dissociate tissues. This becomes even more challenging when working with tissues under inflammatory conditions where cells have been exposed to a harmful and often cytotoxic environment. Some studies have already developed protocols to obtain epithelial and stromal cells from the same tissues (Smillie et al., 2019; Elmentaite et al., 2020); however, there is variability across different studies and laboratories. Our goal was to establish a detailed and easy-to-follow protocol to isolate simultaneously epithelial and stromal cells from mouse and human colon samples under inflammatory conditions. For the mouse protocol, we used the common dextran sodium sulfate (DSS) model of intestinal damage; for the human, we optimized the protocol for processing cryo-preserved colon biopsies from patients with Crohn’s disease. We validated the protocols using downstream flow cytometry and single-cell RNA sequencing analyses and demonstrated that both recover the main cell populations found in the gut mucosa.
Protocol for processing colon tissues from DSS-treated mice
Materials and reagents
Biological materials
7-week-old female CL57BL/6 mice (Charles River Laboratory) were treated with 3% dextran sodium sulfate (DSS) in water for five consecutive days. Colon tissues were isolated on day 3 after treatment at the peak of the inflammatory response.
Reagents
RPMI (Invitrogen, catalog number: 11875119)
Advanced DMEM/F-12 (Gibco, catalog number: 12-634-028)
HBSS (Cytiva, catalog number: SH3058801)
HEPES (Invitrogen, catalog number: 15630080)
GlutaMAX (Thermo Fisher Scientific, catalog number: 35050061)
Pen/Strep (Invitrogen, catalog number: 15140122)
EDTA (0.5 M, pH 8.0) (Invitrogen, catalog number: AM9260G)
DNase I (Roche, catalog number: 10104159001)
Liberase TM (Sigma, catalog number: 05401127001)
TrypLE (Invitrogen, catalog number: 12605010)
Dextran sodium sulfate (DSS) (Alfa Aesar, catalog number: J62101-22)
Trypan Blue solution (Gibco, catalog number: 15250061)
Flow cytometry antibodies:
LIVE/DEADTM Fixable Aqua Dead Cell Stain kit (Thermo Fisher Scientific, catalog number: L34957)
BV421 Rat anti-mouse CD31 (BD Horizon, catalog number: 562939, clone: MEC 13.3)
APC Anti-mouse CD326 (EP-CAM) (BioLegend, catalog number: 118214, clone: G8.8)
PE Anti-mouse Podoplanin (BioLegend, catalog number: 127408, clone: 8.1.1)
FITC Rat anti-mouse CD45 (BD Pharmingen, catalog number: 553080, clone: 30-F11)
Note: Depending on the flow cytometer available, users can use a different color panel design.
Solutions
Epithelial cell solution (see Recipes)
Lamina propria cell solution (see Recipes)
Epithelial wash buffer (see Recipes)
Recipes
Epithelial cell solution
Prepare it fresh and keep it on ice.
Reagent Final concentration Quantity (for one sample) HBSS n/a 11,250 μL HEPES 10 mM 150 μL EDTA (0.5 M) 10 mM 300 μL Pen/Strep 100 U/mL 150 μL FBS 2% 300 μL DNase I 100 μg/mL 150 μL Total n/a 15 mL Lamina propria solution
Prepare it fresh and keep it on ice.
Reagent Final concentration Quantity (for one sample) RPMI n/a 9,600 μL Pen/Strep 100 U/mL 100 μL Liberase TM 100 μg/mL 200 μL DNase I 100 μg/mL 100 μL Total n/a 10 mL Epithelial wash buffer
Once prepared can be stored at 4 °C for one month.
Reagent Final concentration Quantity (for one sample) Adv-DMEM F12 n/a 4,900 μL HEPES 10 mM 500 μL GlutaMAX 10 mM 500 μL Total n/a 50 mL
Laboratory supplies
15 mL centrifuge tube (CELLTREAT, catalog number: 30411B)
50 mL centrifuge tube (CELLTREAT, catalog number: 229430)
100 mm × 15 mm Petri dishes (CELLTREAT, catalog number: 229693)
100 μm cell strainer (Falcon, catalog number: 352360)
40 μm cell strainer (Falcon, catalog number: 352340)
Gavage needle (Gavage Needle, catalog number: AFN2025S)
Frosted microscope slides (Fisher Scientific, catalog number: 12-550-343)
5 mL Polystyrene round-bottom tube with cell-strainer cap (Falcon, catalog number: 352235)
Filter pipette tips (1,000 μL) (Genesee Scientific, catalog number: 24-430)
Serological pipets (10 and 5 mL) (Genesee Scientific, catalog number: 12-102, 12-104)
Equipment
Surgical scissors
Forceps
Centrifuge 5810 R (Eppendorf)
Countess 3 (Invitrogen)
Water bath (Fisher Scientific, Isotemp GPD 10)
Software and datasets
FlowJo v10.8.1
GraphPad Prism9
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Morral, C., Ghinnagow, R., Karakasheva, T., Zhou, Y., Thadi, A., Li, N., Yoshor, B., Soto, G. E., Chen, C. H., Aleynick, D., Weinbrom, S., Fulton, M., Uzun, Y., Bewtra, M., Kelsen, J. R., Lengner, C. J., Tan, K., Minn, A. J. and Hamilton, K. E. (2023). Isolation of Epithelial and Stromal Cells from Colon Tissues in Homeostasis and Under Inflammatory Conditions. Bio-protocol 13(18): e4825. DOI: 10.21769/BioProtoc.4825.
分类
细胞生物学 > 单细胞分析 > 流式细胞术
分子生物学 > RNA > RNA 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link