- EN - English
- CN - 中文
Differentiation of Bone Marrow Monocytes into Alveolar Macrophages-like Cells through Co-culture with Lung Epithelial Cells and Group 2 Innate Lymphoid Cells
通过与肺上皮细胞和第 2 组先天淋巴细胞共培养将骨髓单核细胞分化为肺泡巨噬细胞样细胞
(*contributed equally to this work) 发布: 2023年09月20日第13卷第18期 DOI: 10.21769/BioProtoc.4818 浏览次数: 2457
评审: Rajesh RanjanDebashis DuttaThirupugal Govindarajan
Abstract
During life, the embryonic alveolar macrophage (AM) population undergoes successive waves of depletion and replenishment in response to infectious and inflammatory episodes. While resident AMs are traditionally described as from embryonic origin, their ontogeny following inflammation or infection is much more complex. Indeed, it appears that the contribution of monocytes (MOs) to the AM pool is variable and depends on the type of inflammation, its severity, and the signals released in the microenvironment of the pulmonary niche (peripheral imprinting) and/or in the bone marrow (central imprinting). Deciphering the cellular and molecular mechanisms regulating the differentiation of MOs into AMs remains an area of intense investigation, as this could potentially explain part of the inter-individual susceptibility to respiratory immunopathologies. Here, we detail a relevant ex vivo co-culture model to investigate how lung epithelial cells (ECs) and group 2 lung innate lymphoid cells (ILC2s) contribute to the differentiation of recruited MOs into AMs. Interestingly, the presence of lung ILC2s and ECs provides the necessary niche signals to ensure the differentiation of bone marrow MOs into AMs, thus establishing an accessible model to study the underlying mechanisms following different infection or inflammation processes.
Key features
• Ex vivo co-culture model of the alveolar niche.
• Deciphering the particular niche signals underlying the differentiation of MO into AMs and their functional polarization.
Graphical overview
This protocol described the isolation of bone marrow monocytes (MOs), lung epithelial cells (ECs), and lung group 2 lung innate lymphoid cells (ILC2s) and the ex vivo co-culture of these cells to drive the differentiation of bone marrow MOs into alveolar macrophages (AMs).
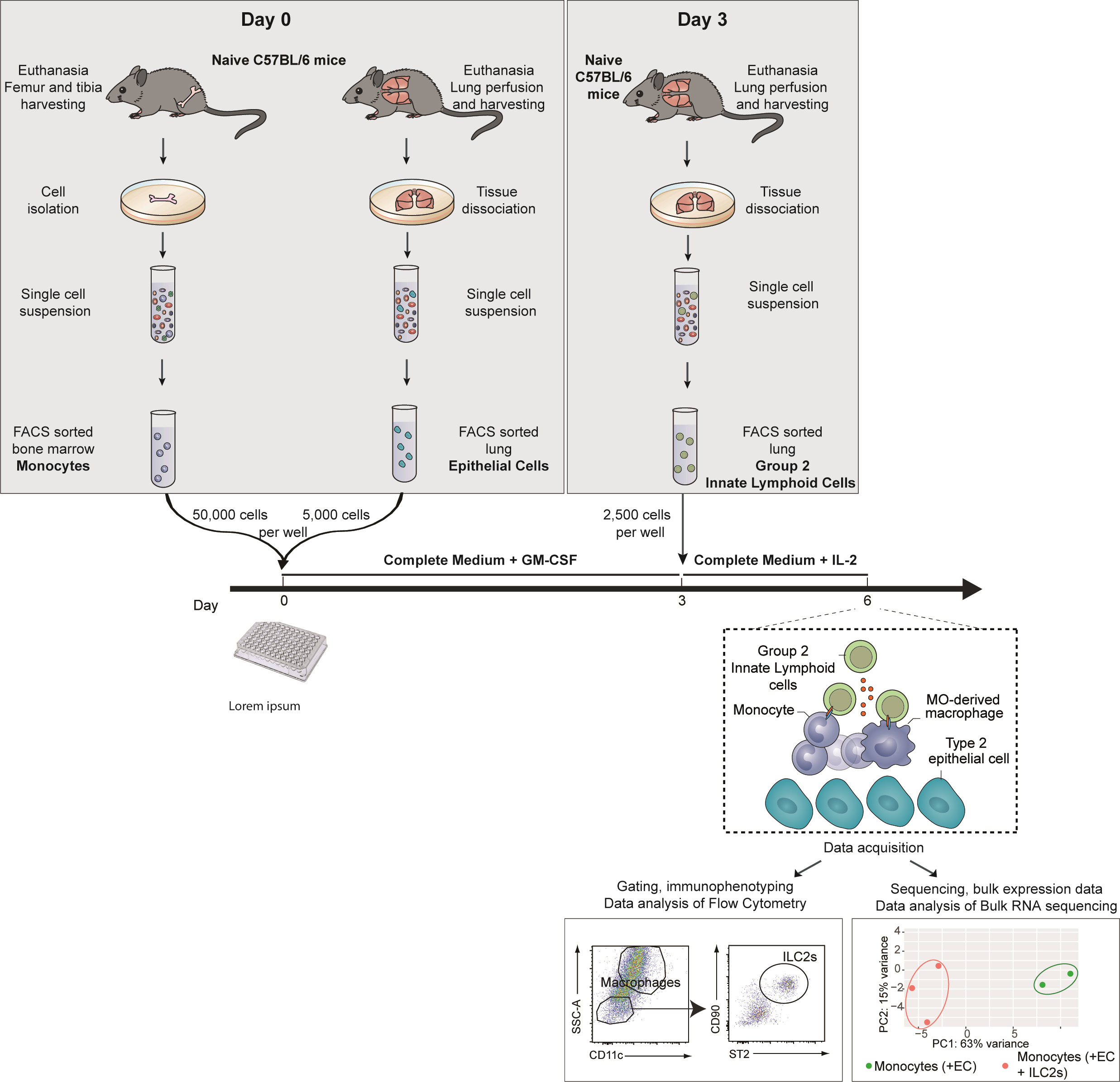
This co-culture experiment is composed of three steps (Graphical overview):
1. Identification and FACS-sorting of ECs and MOs isolated from the lung and the bone marrow of naive mice, respectively.
2. Culture of these ECs and bone marrow MOs for three days.
3. Addition of ILC2s isolated from the lung of naïve mice or mice subjected to a treatment/infection of interest.
Background
Innate lymphoid cells (ILCs) were firstly described in 2010 and have since been described as important regulators of both inflammation and homeostasis throughout the body (Moro et al., 2010; Ebbo et al., 2017). They affect the function of their neighboring cells through the cytokines they produce and also via direct cell–cell interactions (Spits and Di Santo, 2011; Oliphant et al., 2014). Their role in modulating alveolar macrophage (AMs) polarization has been described in early life through the IL-33/ILC2/IL-13 activation cascade, which confers an M2 phenotype to AMs (Saluzzo et al., 2017). In the context of a persistent viral infection, we recently highlighted the essential role of ILC2s as lung niche cells that imprint the tissue-specific identity of monocytes (MOs)-derived AMs and shape their function (Loos et al., 2023).
The protocol presented here details the optimal settings to recapitulate an alveolar niche ex vivo allowing the differentiation of MOs into AMs. In particular, this protocol describes the experimental conditions for co-culture of epithelial cells (ECs) and bone marrow MOs, with group 2 lung innate lymphoid cells (ILC2s) isolated from mouse lung. In the related publication, we showed that bone marrow MOs differentiate in vitro into AM-like cells in the presence of ILC2s and ECs, as evidenced by the expression of genes related to macrophage differentiation and activation (Csf1, PPARg, and Il4ra) (Loos et al., 2023). In addition, we also showed the long-term effect of lung gammaherpesvirus infection on ILC2s and the subsequent consequences on ILC2-mediated differentiation of MOs into AMs. In particular, we highlighted the key role of granulocyte-macrophage colony-stimulating factor (GM-CSF) produced by ILC2s in the differentiation of MO-AMs. The main advantage of this protocol is that it reduces the cell types involved in the differentiation of MO-AMs to a number of selected candidates that we can study ex vivo. This approach therefore allows to test the influence of either environmental conditions (such as viruses or pollutants) or defined mediators in a very controlled environment with every other condition being equal. Subsequently, the differentiation of MOs into AM-like cells can be analyzed classically by multiparametric flow cytometry or by transcriptomic analyses such as bulk or single-cell RNA sequencing. Besides, modifications in intracellular metabolism driving epigenetic reprogramming could be investigated via metabolic (Cedex BioAnalyzer) and epigenetic analysis (ATAC-seq, CHIP-seq). Subsequently, those differences could be confirmed using specific neutralizing antibodies or metabolic (OXPHOS inhibitor, such as oligomycin, or glycolysis manipulator, such as PKM2-pyruvate) (Surace et al., 2021) or epigenetic (BET bromodomain inhibitor IBET151) inhibitors (Kerscher et al., 2019). Altogether, this protocol presents a method to study the different aspects of the biology of MO-derived AMs under controlled and reproducible experimental conditions.
Materials and reagents
Biological materials
Female C57BL/6 mice (8 weeks old) (Charles River, C57BL/6NCrl, catalog number: 000664)
Reagents
Dulbecco’s modified Eagle medium (DMEM, high glucose, GlutaMAX Supplement, pyruvate) (Thermo Fisher Scientific, Gibco, catalog number: 31966021)
RPMI 1640 medium (Thermo Fisher, catalog number: 11875093)
Minimum essential medium (MEM) (Sigma, catalog number: M0446-500ML)
Bovine serum albumin (BSA) (Sigma, catalog number: A8412)
Fetal bovine serum (FBS) (Gibco, catalog number: 10082147)
L-Glutamine (Corning, catalog number: 25005CI)
Sodium pyruvate (Corning, catalog number: 25-000-CIR)
Penicillin-streptomycin (Gibco, catalog number: 15070063)
β-mercaptoethanol (Sigma-Aldrich, catalog number: M3148)
Phosphate-buffered saline, pH 7.4 (Gibco, catalog number: 10010-023)
EDTA (Corning, catalog number: 46-034-CI)
Dispase (Sigma-Aldrich, catalog number: 42613-33-2)
Liberase research grade (Sigma-Aldrich, catalog number: 5401119001)
DNase (Sigma-Aldrich, catalog number: 11284932001)
GentleMACS C tube (Miltenyi, catalog number: 130-093-237)
Red blood cells (RBC) lysis buffer (Thermo Fisher Scientific, catalog number: 00433357)
MojoSort mouse anti-CD45 nanobeads (BioLegend, catalog number: 480027)
MojoSort mouse anti-APC nanobeads (BioLegend, catalog number: 480072)
Recombinant murine GM-CSF (Peprotech, catalog number: 315-03)
Recombinant murine IL-2 (Peprotech, catalog number: 212-12)
Zombie Aqua Fixable Viability kit (BioLegend, catalog number: 423101)
Sytox Blue Dead Cell stain (Thermo Fisher, catalog number: s34857)
Anti-mouse CD16/32 Fc block clone 93 (BioLegend, catalog number: 101301)
Paraformaldehyde (PFA) (Santa Cruz Biotechnology, catalog number: sc-281692)
Saponin (Sigma-Aldrich, catalog number: 47036)
Sodium azide (NaN3) (Sigma-Aldrich, catalog number: S2002)
TRIzol reagent (Thermo Fisher, catalog number: 15596026)
Flow cytometry antibodies
Antibodies for bone marrow MO enrichment (bead-based negative selection) and sorting:
Negative selection
i. Anti-mouse B220/CD45R APC (clone RA3-6B2) (BioLegend, catalog number: 103212)
ii. Anti-mouse Ly6G APC (clone 1A8) (BioLegend, catalog number: 127613)
Flow cytometry mastermix staining
i. Anti-mouse CD11b BV711 (clone M1/70) (BD Biosciences, catalog number: 563168)
ii. Anti-mouse Ly6G APC-Cy7 (clone 1A8) (BD Biosciences catalog number: 560600)
iii. Anti-mouse Ly6C BV785 (clone HK1.4) (BioLegend, catalog number: 128041)
iv. Anti-mouse I-A/I-E PE/Cyanine7 (clone M5/114.15.2) (BioLegend, catalog number: 107630)
v. Anti-mouse CD3e FITC (clone 145-2C11) (BioLegend, catalog number: 100306)
Antibodies for lung EC enrichment (CD45 bead-based negative selection) and sorting:
Flow cytometry mastermix staining
i. Anti-mouse CD45 PE/Cyanine7 (clone 30-F11) (BioLegend, catalog number: 103114)
ii. Anti-mouse EpCAM FITC (clone G8.8) (BioLegend, catalog number: 118207)
iii. Anti-mouse CD31 PE (clone 390) (BioLegend, catalog number: 102407)
Antibodies for lung ILC2 enrichment (bead-based negative selection) and sorting:
Negative selection
i. Anti-mouse B220/CD45R APC (clone RA3-6B2) (BioLegend, catalog number: 103212)
ii. Anti-mouse CD11c APC (clone N418) (BioLegend, catalog number: 117310)
iii. Anti-mouse CD3e APC (clone 145-2C11) (BioLegend, catalog number: 100312)
iv. Anti-mouse CD4 APC (clone RM 4-5) (BioLegend, catalog number: 100515)
v. Anti-mouse CD49b APC (clone DX5) (BioLegend, catalog number: 108910)
vi. Anti-mouse CD5 APC (clone 53-7.3) (BioLegend, catalog number: 100626)
vii. Anti-mouse CD8a APC (clone 53-6.7) (BioLegend, catalog number: 100711)
viii. Anti-mouse F4/80 APC (clone BM8) (BioLegend, catalog number: 123116)
ix. Anti-mouse FcϵRIα APC (clone MAR-1) (BioLegend, catalog number: 134316)
x. Anti-mouse Gr-1 APC (clone RB6-8C5) (BioLegend, catalog number: 108412)
xi. Anti-mouse Siglec-F APC (clone E50-2440) (BioLegend, catalog number: 155507)
Flow cytometry mastermix staining
i. Anti-mouse CD45 PE/Cyanine7 (clone 30-F11) (BioLegend, catalog number: 103114)
ii. Anti-mouse B220/CD45R APC (clone RA3-6B2) (BioLegend, catalog number: 103212)
iii. Anti-mouse CD11c APC (clone N418) (BioLegend, catalog number: 117310)
iv. Anti-mouse CD3e APC (clone 145-2C11) (BioLegend, catalog number: 100312)
v. Anti-mouse CD4 APC (clone RM 4-5) (BioLegend, catalog number: 100515
vi. Anti-mouse CD49b APC (clone DX5) (BioLegend, catalog number: 108910
vii. Anti-mouse CD5 APC (clone 53-7.3) (BioLegend, catalog number: 100626
viii. Anti-mouse CD8a APC (clone 53-6.7) (BioLegend, catalog number: 100711
ix. Anti-mouse F4/80 APC (clone BM8) (BioLegend, catalog number: 123116)
x. Anti-mouse FcϵRIα APC (clone MAR-1) (BioLegend, catalog number: 134316)
xi. Anti-mouse Gr-1 APC (clone RB6-8C5) (BioLegend, catalog number: 108412)
xii. Anti-mouse Siglec-F APC (clone E50-2440) (BioLegend, catalog number: 155507)
xiii. Anti-mouse CD25 Alexa Fluor 700 (clone PC61) (BioLegend, catalog number: 102024)
xiv. Anti-mouse ST2 PE (clone DIH9) (BioLegend, catalog number: 145304)
xv. Anti-mouse CD90.2 BV711 (clone 53-2.1) (BD Biosciences, catalog number: 740647
Solutions
Wash buffer (see Recipes)
MACS buffer (see Recipes)
FACS buffer (see Recipes)
Complete culture medium (see Recipes)
Lung digestion solution (see Recipes)
MO and EC culture medium (see Recipes)
ILC2 culture medium (see Recipes)
Permeabilization buffer (see Recipes)
Intracellular staining buffer (see Recipes)
Recipes
Wash buffer
PBS-EDTA 2 mM with 10% FBS
MACS buffer
PBS containing 0.5% BSA and 2 mM EDTA
Keep buffer cold (2–8 °C)
FACS buffer
PBS containing 0.5% BSA, 2 mM EDTA, 2 mM sodium azide (NaN3)
Complete culture medium
RPMI medium supplemented with 10% FBS, 1% MEM, 100 U/mL penicillin, 100 U/mL streptomycin, 2 mM L-glutamine, 50 μM of β-mercaptoethanol
Lung digestion solution
Freshly prepare 1.5 mL/sample of RPMI containing 50 μg/mL Liberase and 100 μg/mL DNase I
MO and EC culture medium
RPMI medium supplemented with 10% FBS, 1% MEM, 100 U/mL penicillin, 100 U/mL streptomycin, 2 mM L-glutamine, 50 μM of β-mercaptoethanol, and 10 ng/mL recombinant murine GM-CSF
ILC2 culture medium
RPMI-1640 medium supplemented with 10% FBS, 1% MEM, 100 U/mL penicillin, 100 U/mL streptomycin, 2 mM L-glutamine, 50 μM of β-mercaptoethanol, and 10 ng/mL recombinant mouse IL-2
Permeabilization buffer
PBS containing 0.1% of saponin
Keep buffer cold and sterile after filtration
Intracellular staining buffer
FACS buffer (see Recipe 3) containing 0.1% of saponin
Laboratory supplies
15 and 50 mL conical centrifuge tubes (Corning, catalog numbers: 352096 and 352070)
Cell culture 60 × 15 mm Petri dishes (Thermo Fisher Scientific, catalog number: 150288)
24-well cell culture plates (Falcon, catalog number: 353047)
96-well round (U) bottom plate (Thermo Scientific, catalog number: 163320)
10 mL syringes (Terumo, catalog number: SS+10ES1)
25 G needles (Terumo, catalog number: AN*2516R1)
Falcon® 70 μm cell strainer (Corning, catalog number: 352350)
LD columns (Miltenyi, catalog number: 130-042-901)
Equipment
Swing-bucket centrifuge (Eppendorf, model: 5810R)
Tabletop centrifuge (Eppendorf, model: 5427R)
Inverted light microscope (NIKON, Eclipse TS100)
Fluorescence-activated cell sorter (BD Biosciences, model: Aria IIIu)
Flow cytometer (BD LSR Fortessa X-20)
Laminar flow hood (FASTER SafeFast Classic)
Scalpel (Swann-Morton 210 mm No.3 ref 0933)
Forceps (Bochem Stainless steel 18/10 ref 1141)
GentleMACS dissociator (Miltenyi)
QuadroMACS Separators (Miltenyi)
Hemocytometer (Marienfeld, 0640710)
Software and datasets
FlowJo v10 (FlowJo LLC)
FACSDiVa software
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Loos, P., Marichal, T., Machiels, B. and Gillet, L. (2023). Differentiation of Bone Marrow Monocytes into Alveolar Macrophages-like Cells through Co-culture with Lung Epithelial Cells and Group 2 Innate Lymphoid Cells. Bio-protocol 13(18): e4818. DOI: 10.21769/BioProtoc.4818.
- Loos, P., Baiwir, J., Maquet, C., Javaux, J., Sandor, R., Lallemand, F., Marichal, T., Machiels, B. and Gillet, L. (2023). Dampening type 2 properties of group 2 innate lymphoid cells by a gammaherpesvirus infection reprograms alveolar macrophages. Sci. Immunol. 8(80): eabl9041.
分类
免疫学 > 免疫细胞分离 > 巨噬细胞
细胞生物学 > 细胞分离和培养 > 共培养
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link