- EN - English
- CN - 中文
Spatial Centrosome Proteomic Profiling of Human iPSC-derived Neural Cells
人 iPSC 衍生神经细胞的空间中心体蛋白质组学分析
(*contributed equally to this work) 发布: 2023年09月05日第13卷第17期 DOI: 10.21769/BioProtoc.4812 浏览次数: 2542
评审: Alessandro DidonnaAbraam YakoubYiqun Yu
Abstract
The centrosome governs many pan-cellular processes including cell division, migration, and cilium formation. However, very little is known about its cell type-specific protein composition and the sub-organellar domains where these protein interactions take place. Here, we outline a protocol for the spatial interrogation of the centrosome proteome in human cells, such as those differentiated from induced pluripotent stem cells (iPSCs), through co-immunoprecipitation of protein complexes around selected baits that are known to reside at different structural parts of the centrosome, followed by mass spectrometry. The protocol describes expansion and differentiation of human iPSCs to dorsal forebrain neural progenitors and cortical projection neurons, harvesting and lysis of cells for protein isolation, co-immunoprecipitation with antibodies against selected bait proteins, preparation for mass spectrometry, processing the mass spectrometry output files using MaxQuant software, and statistical analysis using Perseus software to identify the enriched proteins by each bait. Given the large number of cells needed for the isolation of centrosome proteins, this protocol can be scaled up or down by modifying the number of bait proteins and can also be carried out in batches. It can potentially be adapted for other cell types, organelles, and species as well.
Graphical overview

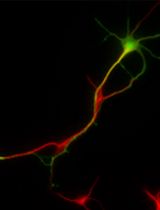
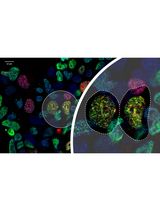
An overview of the protocol for analyzing the spatial protein composition of the centrosome in human induced pluripotent stem cell (iPSC)-derived neural cells. ① Human iPSCs are expanded, which serve as the starting cell population for the neural induction (Sections A, B, and C in Procedure). ② Neurons are induced and differentiated for 40 days (Section D in Procedure), in at least four biological replicates. ③ Total protein is isolated either at 15th or 40th day of differentiation, for neural stem cells and neurons, respectively (Sections E and F in Procedure). ④ Selected bait proteins are immunoprecipitated using the respective antibodies (Sections G and H in Procedure). ⑤ Co-immunoprecipitated samples are analyzed with mass spectrometry (Section I in Procedure). ⑥ Mass spectrometry output (.RAW) files are processed using MaxQuant software to calculate intensities (Section A in Data analysis). ⑦ The resulting data are pre-processed, filtered, and statistically analyzed using Perseus and R software (Sections B and C in Data analysis) ⑧ Further analysis is done using software or web tools such as Cytoscape or STRING to gain biological insights (Sections D and E in Data analysis).
Background
The centrosome is a multifunctional organelle that is known primarily for its microtubule organizing functions, as well as roles related to cell cycle, cell polarity, and migration (Bornens, 2021). Embedded in the pericentriolar material, the centrosome is a highly ordered structure whose core is made of a pair of orthogonally oriented centrioles, each with nine triplets of microtubules with differing maturity—named the mother and daughter centrioles (Fu et al., 2015).
The centrioles exhibit structural differences such as the presence or absence of subdistal and distal appendages, which also facilitate their functional distinction. For example, the more mature mother centriole has a higher capacity to anchor microtubules via these appendages, which are also used in membrane docking—a prerequisite in cilium formation (Bornens, 2002; Brugués et al., 2012; Paridaen et al., 2013; Tanos et al., 2013). The pericentriolar material surrounding the centrosome also serves important functions such as microtubule nucleation, and also contains important signaling proteins (Woodruff et al., 2014; Loukil et al., 2017; Lin et al., 2022).
Although traditionally assumed to be largely composed of a set of homogenous proteins across cell types that support its generic functions, several centrosomal proteins have been identified based on candidate studies that reveal the presence of cell type–specific dynamic relationships (Camargo Ortega and Götz, 2022). For example, the centrosomal protein formerly named AT-hook-containing transcription factor (AKNA) differentially localizes to the subdistal appendages of the centrosomes in neural stem cells and neurons to govern specific processes associated with brain development as well as cell fate determination (Camargo Ortega et al., 2019). NINEIN is another dynamically associated centrosome protein, whose loss from the subdistal appendages in neurons leads to the loss of centrosomal microtubule organizing functions (Wang et al., 2009; Shinohara et al., 2013; Zhang et al., 2016).
Although centrosome proteomes have been cataloged for cancer cells and Drosophila (Andersen et al., 2003; Sauer et al., 2005; Müller et al., 2010; Gheiratmand et al., 2019), these few dynamically identified proteins call for more widespread analysis of interacting partners among a range of cell types. The large amount of cellular material needed for centrosome isolation via fractionation and its inconsistent fractionation behavior prohibits, however, large-scale analyses and does not inform on the location of the newly identified interacting partners on the centrosome. Bio-ID facilitates such proximity labeling, but is compounded by overexpressing a tagged fusion protein, potentially impacting endogenous centrosome functions that might be sensitive to its levels within a cell (Sears et al., 2019). Proteins identified through Bio-ID protocols may also not biochemically interact but rather just localize near the protein/structure of interest (Rattray and Foster, 2019).
We recently applied co-immunoprecipitation of core endogenous centrosomal proteins coupled with mass-spectrometry to outline an effective alternative to both fractionation and tagging strategies (O’Neill et al., 2022). Using core centrosomal proteins with overlapping but non-redundant positions within this organelle, we could resolve spatial interaction networks across the cell types investigated (human dorsal neural stem cells and neurons), identifying the centrosome as a hub for RNA-binding proteins that also explain how ubiquitous proteins can cause organ-specific disease phenotypes when mutated. The use of the microtubule depolymerizing agent, nocodazole, offered an additional mechanism to identify the microtubule-dependent and -independent interacting partners within the cell types of interest (O’Neill et al., 2022).
Here, we outline in detail this method for interrogating the spatial centrosome proteome in human cells. We begin by describing the specific culture of the neural cells starting from human induced pluripotent stem cells (iPSCs), followed by the co-immunoprecipitation. We end with a detailed description of the analysis of proteomic data resulting from the previous steps. Adopting this strategy across a range of cell types can potentially offer further insights into the spatial organization and heterogeneity of the centrosome, extending the functional repertoire of this organelle. The approach can be further adapted to different sub-cellular targets or tissues, as well as other cell types including primary cells isolated from different species.
Materials and reagents
Human induced pluripotent stem cell lines, HMGU1 (hPSCreg: ISFi001-A) and HMGU12 (hPSCreg: ISFi002-A), obtained from the Helmholtz Center Munich iPSC Core Facility. The source of the cells is BJ fibroblasts (ATCC CRL-2522)
mTeSRTM 1 culture media (basal medium and 5× supplement) (StemCell Technologies, catalog number: 05850)
DMEM/F-12, GlutaMAXTM supplement (GibcoTM, catalog number: 10565018)
Neurobasal medium (1×) [-] L-Glutamine (GibcoTM, catalog number: 21103-049)
B-27 supplement with vitamin A (50×) (GibcoTM, catalog number: 17504044)
GlutaMAXTM supplement (100×) (GibcoTM, catalog number: 35050061)
Penicillin-Streptomycin (GibcoTM, catalog number: 15140122)
N2 supplement (100×) (GibcoTM, catalog number: 17502048)
Non-essential amino acids (100×) (GibcoTM, catalog number: 11140050)
2-mercaptoethanol (50 mM) (GibcoTM, catalog number: 31350010)
Insulin, human (10 mg/mL) (Sigma-Aldrich, catalog number: I9278-5ML)
Collagenase type IV (1 mg/mL) (StemCell Technologies, catalog number: 07909)
StemProTM AccutaseTM (GibcoTM, catalog number: A1110501)
Geltrex Matrix, reduced growth factor (GibcoTM, catalog number: A1413202)
Matrigel® growth factor reduced (Corning®, catalog number: 354230)
Laminin from Engelbreth-Holm-Swarm murine sarcoma basement membrane (Sigma-Aldrich, catalog number: L2020-1MG)
Poly-L-Ornithine hydrobromide (Sigma-Aldrich, catalog number: P3655-500MG)
Dorsomorphin (Sigma-Aldrich, catalog number: P5499-5MG)
SB431542 hydrate (Sigma-Aldrich, catalog number: S4317-5MG)
Recombinant human FGF-basic (Peprotech, catalog number: 100-18B)
ROCK inhibitor (Y-27632), 5 mg (StemCell Technologies, catalog number: 72304)
Nocodazole (Sigma-Aldrich, catalog number: M1404)
Dimethyl sulfoxide (DMSO), sterile-filtered (Sigma-Aldrich, catalog number: D2438)
Phosphate buffered saline (PBS) (10×) (GibcoTM, catalog number: 70011044)
Sterile water (Braun, catalog number: 0082423E)
MilliQ water (Merck) or distilled water (any brand)
Media filter: Filtropur V25, 250 mL, 0.2 μm (Sarstedt, catalog number: 83.3940.001)
Media filter: Filtropur V50, 500 mL, 0.2 μm (Sarstedt, catalog number: 83.3941.001)
Tris-base [NH2C(CH2OH)3] (Millipore, catalog number: 648310)
Sodium chloride (NaCl) (Sigma, catalog number: S3014)
Ethylenediaminetetraacetic acid tetrasodium salt (EDTA) (Sigma, catalog number: E6758)
Ethylene glycol-bis(2-aminoethylether)-tetraacetic acid (EGTA) (Sigma, catalog number: E4378)
Triton X-100 (Bio-Rad, catalog number: 1610407)
Sodium dodecyl sulfate (SDS) (Sigma, catalog number: L3771)
2-mercaptoethanol, molecular biology grade (Sigma-Aldrich, catalog number: M3148-25ML)
cOmplete, Mini Protease Inhibitor Cocktail (Roche, catalog number: 11836153001)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A2153)
Protein assay reagent A (Bio-Rad, catalog number: 5000113)
Protein assay reagent B (Bio-Rad, catalog number: 5000114)
Protein assay reagent S (Bio-Rad, catalog number: 5000115)
DynabeadsTM protein A (Invitrogen, catalog number: 10002D)
DynabeadsTM protein G (Invitrogen, catalog number: 10004D)
Micropipette tips (any brand)
6-well cell culture dishes (Falcon®, catalog number: 353046)
10 cm cell culture dishes (Falcon®, catalog number: 353003)
Cell scrapers (TPPTM, catalog number: 99002) (Sarstedt, catalog number: 83.3951)
50 mL Falcon tubes, 15 mL Falcon tubes (any brand)
Protein LoBind® Tubes, 1.5 and 2 mL (Eppendorf, catalog numbers: 0030108116 and 0030108132)
Protein LoBind® Tubes, 50 mL (Eppendorf, catalog number: 0030122240)
5, 10, and 25 mL serological pipettes (any brand)
Rabbit polyclonal anti-CDK5RAP2 (Sigma-Aldrich, catalog number: 06-1398)
Mouse polyclonal anti-CNTROB (Abcam, catalog number: ab70448)
Rabbit polyclonal anti-CP110 (Abcam, catalog number: ab99338)
Mouse monoclonal anti-CEP170 (InvitrogenTM, catalog number: 72-413-1/41-3200)
Rabbit polyclonal anti-CEP192 (Novus Biological, catalog number: NBP1-28718)
Rabbit polyclonal anti-CEP152 (Merck Millipore, catalog number: ABE1856)
Rabbit polyclonal anti-CEP63 (Merck Millipore, catalog number: 06-1292)
Rabbit polyclonal anti-CEP135 (Antibodies online, catalog number: ABIN2801434)
Rabbit polyclonal anti-POC5 (Novus Biological, catalog number: NBP1-78741)
Rabbit polyclonal anti-ODF2 (Abcam, catalog number: ab43840)
ROCK inhibitor (10 mM) (see Recipes)
Dorsomorphin (5 mM) (see Recipes)
SB431542 (10 mM) (see Recipes)
Poly-L-Ornithine, 10 mg/mL (see Recipes)
Neural maintenance medium (N3) (see Recipes)
Neural induction medium (N3+SMADi) (see Recipes)
Nocodazole (3.3 mM) (see Recipes)
2 M NaCl (see Recipes)
0.5 M EDTA, pH 7.5 (see Recipes)
0.5 M EGTA, pH 7.6 (see Recipes)
1 M Tris-HCl, pH 7.5 (see Recipes)
0.5 M Tris-HCl, pH 6.8 (see Recipes)
10% Triton X-100 (v/v) (see Recipes)
10% SDS (w/v) (see Recipes)
IP/wash buffer A (see Recipes)
Wash buffer B (see Recipes)
Protein standard (BSA) (64 mg/mL) (see Recipes)
1× Laemmli buffer (see Recipes)
Equipment
Inverted microscope (Leica, DM IL LED)
Neubauer improved cell counting chamber (Assistent, catalog number: 40442702)
Humidified cell culture incubator (any brand)
Brand Accu-jet pro pipette controller (Pro-lab, catalog number: 26330)
Microliter pipettes (Gilson®, P1000, catalog number: 10387322; P200, catalog number: 10327282; P20, catalog number: 10082012; P2, catalog number: 10635313)
Ice bucket (any brand)
Spectrophotometer or microplate reader (any brand with 750 nm wavelength)
DynaMag-2 magnet (Thermo Fisher Scientific, catalog number: 12321D)
1.5 mL vertical tube rotator (Stuart, catalog number: SB3)
Microcentrifuge with refrigeration (Thermo Fisher Scientific, catalog number: 75002446)
Microcentrifuge with refrigeration (Thermo Fisher Scientific, catalog number: 75004510)
Chemical fume hood (any brand)
Heating block (any brand with 95 °C capacity)
QExactive HF mass spectrometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA)
Software
MaxQuant: https://www.maxquant.org/
Perseus: https://www.maxquant.org/perseus/
R Statistical Software: https://www.r-project.org/
Microsoft Excel: https://www.microsoft.com/de-de/microsoft-365/excel
Web resources
Ensembl Genome Browser: http://www.ensembl.org/index.html
Proteomics Identification Database (PRIDE): https://www.ebi.ac.uk/pride/
STRING Database: https://string-db.org/
Perseus documentation: http://www.coxdocs.org/doku.php?id=:perseus:start
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Uzbas, F. and O'Neill, A. (2023). Spatial Centrosome Proteomic Profiling of Human iPSC-derived Neural Cells. Bio-protocol 13(17): e4812. DOI: 10.21769/BioProtoc.4812.
- O’Neill, A. C., Uzbas, F., Antognolli, G., Merino, F., Draganova, K., Jäck, A., Zhang, S., Pedini, G., Schessner, J. P., Cramer, K., et al. (2022). Spatial centrosome proteome of human neural cells uncovers disease-relevant heterogeneity. Science 376(6599): eabf9088.
分类
神经科学 > 发育 > 神经元
发育生物学 > 细胞生长和命运决定 > 分化
分子生物学 > 蛋白质 > 蛋白质-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link