- EN - English
- CN - 中文
Endoplasmic Reticulum Isolation: An Optimized Approach into Cells and Mouse Liver Fractionation
内质网分离:细胞和小鼠肝脏分离的优化方法
(*contributed equally to this work) 发布: 2023年09月05日第13卷第17期 DOI: 10.21769/BioProtoc.4803 浏览次数: 3487
评审: Davide BottaAmberley D. StephensMichael D Schultz
Abstract
The subfractionation of the endoplasmic reticulum (ER) is a widely used technique in cell biology. However, current protocols present limitations such as low yield, the use of large number of dishes, and contamination with other organelles. Here, we describe an improved method for ER subfractionation that solves other reported methods' main limitations of being time consuming and requiring less starting material. Our protocol involves a combination of different centrifugations and special buffer incubations as well as a fine-tuned method for homogenization followed by western blotting to confirm the purity of the fractions. This protocol contains a method to extract clean ER samples from cells using only five (150 mm) dishes instead of over 50 plates needed in other protocols. In addition, in this article we not only propose a new cell fractionation approach but also an optimized method to isolate pure ER fractions from one mouse liver instead of three, which are commonly used in other protocols. The protocols described here are optimized for time efficiency and designed for seamless execution in any laboratory, eliminating the need for special/patented reagents.
Key features
• Subcellular fractionation from cells and mouse liver.
• Uses only five dishes (150 mm) or one mouse liver to extract highly enriched endoplasmic reticulum without mitochondrial-associated membrane contamination.
• These protocols require the use of ultracentrifuges, dounce homogenizers, and/or Teflon Potter Elvehjem.
• As a result, highly enriched/clean samples are obtained.
Graphical overview
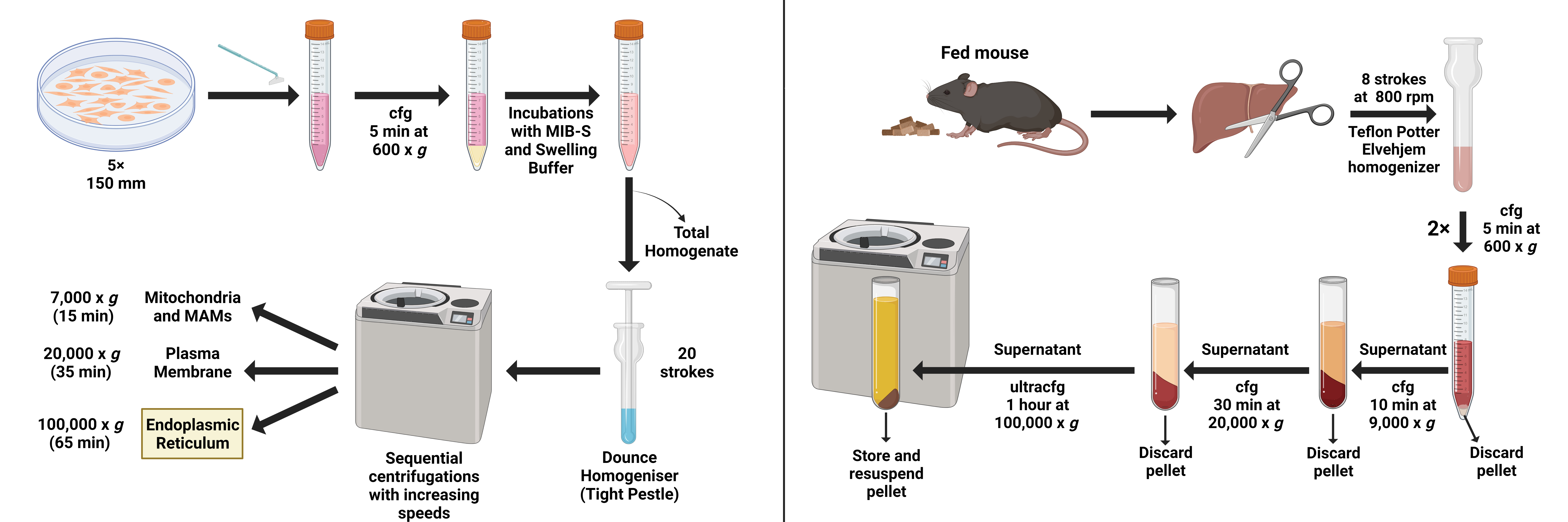
Background
The endoplasmic reticulum (ER) is a continuous membrane system that forms a series of flattened sacs within the cytoplasm of eukaryotic cells and serves multiple functions. Some of its most relevant roles include the synthesis, folding, modification, and transport of proteins (Schwarz and Blower, 2016). The ER was first observed by electron microscopy in 1945 by Porter and Claude (Porter et al., 1945), but its isolation as a distinct organelle was not achieved until 1959 by Palade and Siekevitz (Siekevitz and Palade, 1959). In this study, authors used differential centrifugation to separate ER membranes from other cellular components. This method was later refined by adding density gradient centrifugations to obtain cleaner (non-other contaminating organelles marker) fractions (Lee et al., 2010).
The isolation of the ER has facilitated and enhanced the understanding of its structure, function, and interactions with other organelles. Some of these studies revealed that the ER forms contact sites with other membranes, such as plasma membrane–associated membranes (PAMs) (Suski et al., 2014) and mitochondria-associated membranes (MAMs) (Missiroli et al., 2018), which mediate transport of proteins, lipids, and metabolites (Ventura and Isabel Hernández-Alvarez, 2022).
However, despite being a highly common technique, the isolation of ER has some limitations. Firstly, it requires a large amount of starting material (usually several grams of tissue or cell number) and takes a long time to complete (6 h in cells because of the time needed to recover the starting material from the plates). Secondly, existing protocols may not be suitable for all tissues or cell types that have different membrane properties or distributions. Thirdly, it is quite difficult to obtain pure samples, and thus achieving highly enriched fractions can be challenging.
Most already existing methods are able to isolate ER indirectly, but only a very small number of protocols are dedicated to the extraction of this particular organelle (Croze and Morré, 1984). This scenario can be found in some of the published protocols that use mouse liver, such as the ones described by Wieckowski et al. (2009) or Suski et al. (2014), where they propose a method to isolate specific areas of the ER, such as MAMs and PAMs, respectively. This fact can also be a limiting factor.
In regard to HeLa cells, no protocols were found to specifically extract the ER as well. Moreover, some protocols that work with similar adherent cells require a large number of dishes (over 50 dishes) (Wieckowski et al., 2009; Williamson et al., 2015), which complicates the isolation and the previous cell culture work. Additionally, a similar situation happens with subcellular fractionations that use liver tissue. According to published protocols, fractionations of liver tissue are performed using rat liver rather than mouse liver, and they usually require from 8 to 10 g of tissue (Suski et al., 2014). When extrapolated to a mouse model, five or six animals for every subfractionation are needed.
Since our laboratory is mainly focused on the study of mitochondrial dynamics, ER–mitochondria contacts, and its relationship with metabolic diseases (Hernández-Alvarez et al., 2019), a protocol that overcame some of these limitations was necessary. Hence, in this article, we provide an optimized and upgraded version of already existing protocols for ER isolation achieving a high throughput using the minimum amount of starting sample. Furthermore, the protocols described below are optimized for time efficiency and designed for seamless execution in any laboratory, eliminating the need for special/patented reagents. Other protocols do not report the yield of ER.
Materials and reagents
Biological materials
Mouse liver or cell line of interest
Antibodies
Mouse monoclonal anti-PDI (C-2) (Santa Cruz Biotechnology, catalog number: sc-74551)
Rabbit monoclonal anti-Tom20 (D8T4N) (Cell Signalling Technology, catalog number: 42406)
Mouse monoclonal anti-VDAC1 (B-6) (Santa Cruz Biotechnology, catalog number: 390996)
Mouse monoclonal anti-Tim23 (H-8) (Santa Cruz Biotechnology, catalog number: sc-514463)
Rabbit monoclonal anti-FACL4 (EPR8640) (Abcam, catalog number: ab155282)
Mouse monoclonal anti-Na+/K+ ATPase alpha 3 subunit (Sigma-Aldrich, catalog number: 05-369-25UG)
Reagents
Isolation from cells
HEPES (Merck, Sigma-Aldrich, catalog number: 54461)
D-Sucrose (Thermo Fisher Scientific, Fisher bioreagents, catalog number: BP220-1)
EGTA (Merck, Calbiochem, catalog number: 324626)
KCl (Honeywell, Fluka analytical, catalog number: 31248)
Isolation from mouse liver
D-Mannitol (Merck, Calbiochem, catalog number: 443907)
D-Sucrose (Thermo Fisher Scientific, Fisher bioreagents, catalog number: BP220-1)
Albumin from bovine serum (BSA) (Merck, Sigma-Aldrich, catalog number: A7906)
EGTA (Merck, Calbiochem, catalog number: 324626)
Tris hydrochloride (Tris-HCl) for buffer solutions (PanReac AppliChem, catalog number: A1087)
10× PBS
NaCl (Thermo Fisher Scientific, catalog number: 11916388)
Na2HPO4·2H2O (Thermo Fisher Scientific, catalog number: 15613040)
KH2PO4 (Merck, catalog number: 104873)
KCl (Honeywell, Fluka analytical, catalog number: 31248)
Solutions
Microsome isolation buffer cells (MIB-C) (see Recipes)
Microsome isolation stability buffer (MIB-S) (see Recipes)
5× swelling buffer (SB) (see Recipes)
Microsome isolation buffer liver (MIB-L) (see Recipes)
10× PBS (see Recipes)
Recipes
Microsome isolation buffer liver (MIB-L)
Reagent Final concentration Quantity D-Mannitol 225 mM 0.41 g D-Sucrose 75 mM 0.26 g EGTA (100 mM; pH 8) 0.5 mM 0.05 mL Tris-HCl (1 M, pH 6.5) 30 mM 0.3 mL BSA 0.5% 0.05 g MQH2O n/a Up to 10 mL Total n/a 10 mL Microsome isolation buffer cells (MIB-C)
Reagent Final concentration Quantity Tris-HCl (1 M, pH 6.5) 30 mM 450 μL D-Mannitol 225 mM 0.61 g D-Sucrose 75 mM 0.38 g MQH2O n/a Up to 15 mL Total n/a 15 mL Microsome isolation stability buffer (MIB-S)
Reagent Final concentration Quantity D-Sucrose 0.25 M 1.2 g KCl 25 mM 0.028 g HEPES (1 M) 10 mM 150 μL EGTA (100 mM; pH 8) 1 mM 150 μL MQH2O n/a Up to 15 mL Total n/a 15 mL 5× swelling buffer (SB)
Reagent Final concentration Quantity KCl 125 mM 0.14 g HEPES (1 M) 50 mM 750 μL EGTA (100 mM; pH 8) 5 mM 150 μL MQH2O n/a Up to 15 mL Total n/a 15 mL 10× PBS
Reagent Final concentration Quantity NaCl 1.5 M 400 g Na2HPO4·2H2O 0.1 M 80 g KH2PO4 15 mM 10 g KCl 25 mM 10 g MQH2O n/a up to 5 L Total n/a 5 L
Equipment
Corning® tissue culture–treated culture dishes (150 mm × 25 mm) (Merck, Corning®, catalog number: CLS430599)
Active Motif dounce homogenizer (Fisher Scientific, Active Motif 40401, catalog number: NC0569256)
NuncTM cell scrapers (Thermo ScientificTM, catalog number: 179707PK)
L-90K ultracentrifuge (Beckman coulter, catalog number: 8043-30-1191)
Type 90 Ti fixed-angle titanium rotor (Beckman coulter, catalog number: 355530)
Centrifuge 5810R (Eppendorf, catalog number: 5811000015)
15 mL polypropylene centrifugation tube (Thermo ScientificTM, catalog number: 17627105)
8.9 mL ultracentrifuge tube (Beckman coulter, catalog number: 361660)
Heidolph RZR 2051 control homogenizer (Fisher Scientific, catalog number: FIS13-880-115)
Tissue grinders, Potter Elvehjem type with PTFE pestle for soft tissue, 15 cm3 (Avantor, VWR®, catalog number 432-5041)
Western blotting equipment (Bio-Rad)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Leiro, M., Ventura, R., Rojo, N. and Hernández-Alvarez, M. I. (2023). Endoplasmic Reticulum Isolation: An Optimized Approach into Cells and Mouse Liver Fractionation. Bio-protocol 13(17): e4803. DOI: 10.21769/BioProtoc.4803.
分类
细胞生物学 > 细胞器分离 > 内质网
细胞生物学 > 细胞器分离 > 分级分离
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link