- EN - English
- CN - 中文
Detection of Cytoplasmic and Nuclear Circular RNA via RT-qPCR
RT-qPCR 检测细胞质和细胞核环状 RNA
发布: 2023年09月05日第13卷第17期 DOI: 10.21769/BioProtoc.4798 浏览次数: 3383
评审: Durai SellegounderSalim Gasmi

相关实验方案
用细胞增殖和基因表达测定法从柏氏血矛线虫排泄物和分泌物中筛选具有免疫潜力的分子
Jocelyn Maza-Lopez [...] Carla O. Contreras-Ochoa
2023年06月20日 1316 阅读
Abstract
Circular RNA (circRNA) is an intriguing class of non-coding RNA that exists as a continuous closed loop. With the improvements in high throughput sequencing, biochemical analysis, and bioinformatic algorithms, studies on circRNA expression became abundant in recent years. However, functional studies of circRNA are still limited. Subcellular localization of circRNA may provide some clues in elucidating its biological functions by performing subcellular fractionation assay. Notably, circRNAs that are predominantly found in the cytoplasm are more likely to be involved in post-transcriptional gene regulation, e.g., acting as micoRNA sponge, whereas nuclear-retained circRNAs are predicted to play a role in transcriptional regulation. Subcellular fractionation could help researchers to narrow down and prioritize downstream experiments. The majority of the currently available protocols describe the steps for subcellular fractionation followed by western blot analysis for protein molecules. Here, we present a protocol for the subcellular fractionation of cells to detect circRNA via RT-qPCR with divergent primers. Moreover, detailed steps for the generation of specific circRNAs-enriched cDNA included in this protocol will enhance the amplification and detection of low-abundance circRNAs. This will be useful for researchers studying low-abundance circRNAs.
Key features
• This protocol builds upon the method developed by Gagnon et al. (2014) and extends its application to circRNA study.
• Protocol for amplification of low levels of circRNA expression.
• Analysis takes into consideration the ratio of cytoplasmic RNA concentration to nuclear RNA concentration.
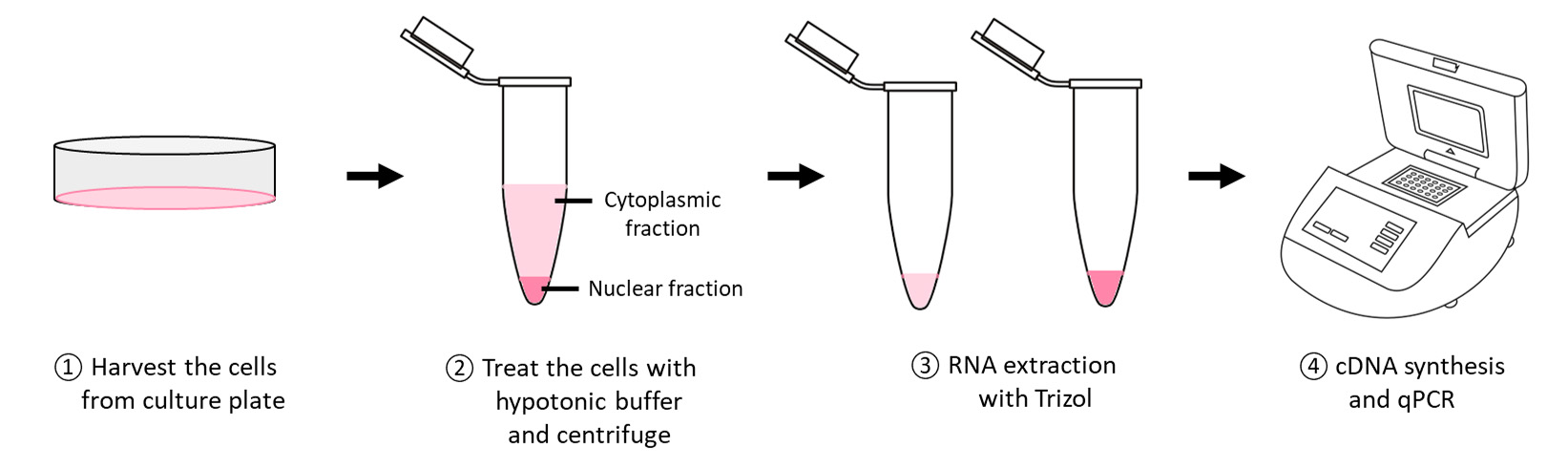
Graphical overview
Background
Circular RNA (circRNA) is an intriguing class of non-coding RNA that is formed through a unique mechanism known as backsplicing, in which the 5′ and 3′ termini are covalently joined (Jeck and Sharpless, 2014). Due to the absence of free termini in the circular structure, circRNA can easily escape from hydrolysis by numerous cellular exonucleases. CircRNAs can be derived from exons, introns, or both, with a great diversity in length. Emerging evidence has shown that circRNAs are involved in many different biological processes and human diseases by functioning as miRNA sponges, RNA-binding protein sponges, transcriptional regulators, mRNA traps, regulators of assembly and transport of cellular proteins, as well as templates for translation.
Several computational methods, such as CIRI2, CIRCexplorer2, and find_circ, have been developed to identify circRNA by employing alignment-based strategies to recognize the back-spliced junction (BSJ), which is a unique feature of circRNAs (Szabo and Salzman, 2016; Cai et al., 2020). To confirm the presence of a circRNA identified in silico, an experimental approach is applied to validate the computationally predicted circRNAs (Zhang et al., 2016).
Reverse-transcription PCR (RT-PCR) is commonly used for the validation of circRNAs identified by RNA sequencing using divergent primers. In this approach, circRNA is converted to cDNA via reverse transcription (RT) with the presence of random hexamer or gene-specific reverse primers. Priming with random hexamers offers flexibility by ensuring RT of all RNA sequences, but gene-specific RT enhances the sensitivity of detecting the target of interest by converting the target of interest—instead of transcribing all RNA in the mix—into cDNA. Priming with gene-specific primers could enhance the detection of circRNA of interest, especially for those present in low abundance (Bustin and Nolan, 2004).
The conventional RT-PCR assay using convergent primers is unable to differentiate circRNAs from their linear transcript when the linear genome is used as template for primer design. Divergent primers are needed for the RT-PCR, as they allow direct detection of BSJ and quantification of circRNA. Divergent primers are a pair of outward-facing primers that allow amplification of circRNAs and not linear mRNA with the same sequence in cDNA and not gDNA (Figure 1). Sanger sequencing is usually performed on the PCR product amplified with the divergent primers to confirm the presence of a BSJ, as to rule out any incorrect amplification or artifact. Quantitative RT-PCR (RT-qPCR) can be used to relatively quantify the expression of circRNA across a panel of samples.
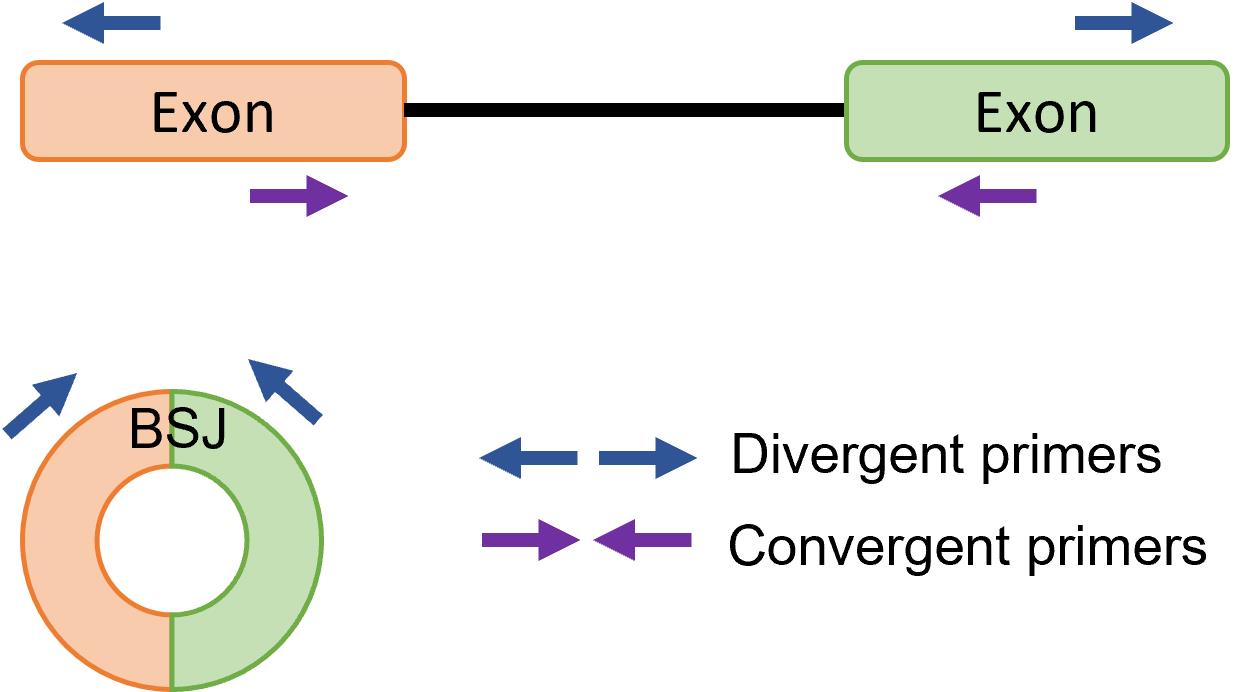
Figure 1. Primer design for circular RNA (circRNA) detection. Divergent primers are a pair of outward-facing primers that allow amplification of circRNA backsplice junction (BSJ), whereas convergent primers are a pair of inward-facing primers that allow amplification of linear mRNA from prepared cDNA.
Combining subcellular fractionation and RT-qPCR with divergent primers may provide some hints on circRNA’s biological functions. For example, circRNAs that are predominantly found in the cytoplasm are more likely to be involved in post-transcriptional gene regulation, whereas nuclear-retained circRNAs are predicted to play a role in transcription regulation. Currently available protocols are mainly for protein work, describing the steps for subcellular fractionation followed by western blot analysis, which is not entirely suitable for RNA work. Instead, this protocol allows the downstream detection of circRNA through RT-qPCR with divergent primers. A detailed protocol for the generation of specific circRNAs-enriched cDNA will also be included to better detect low-abundance circRNAs.
Materials and reagents
Biological material
GM12878 cell line
Reagents
Dulbecco’s phosphate-buffered saline (PBS) (Fisher Scientific, catalog number: BP399-1)
Trypsin-EDTA solution (0.5%) (Gibco, catalog number: 15400054)
Tissue culture media: Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, catalog number: 11875093)
Fetal bovine serum (FBS) (Gibco, catalog number: 10270106)
Penicillin-Streptomycin (Gibco, catalog number: 15140122)
Trypan blue (Alfa Aesar, catalog number: A1860)
Tris-Base (Fisher Scientific, catalog number: BP152-5)
Potassium chloride (KCl) (Fisher Scientific, catalog number: BP366-1)
Magnesium chloride (MgCl2) (Fisher Scientific, catalog number: BP214-500)
DL-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632)
NP-40 (Sigma-Aldrich, catalog number: 9016-45-9)
Ribonucleoside vanadyl complexes (NEB, catalog number: S1402S)
Sodium acetate (Fisher Scientific, catalog number: BP333-500)
Molecular grade ethanol (Merck, catalog number: 64-17-5)
TRIzol (Life Technologies, catalog number: 15-596-018)
Chloroform:isoamyl alcohol (1:24) (Fisher Scientific, catalog number: BP1752I-400)
Molecular grade isopropanol (Fisher Scientific, catalog number: BP2618-1)
RNase-free water (Macherey Nigel, catalog number: 740378)
10× M-MuLV RT buffer (NEB, catalog number: #M0253)
DNase I (NEB, catalog number: #M0303)
M-MuLV reverse transcriptase (NEB, catalog number: #M0253)
RNase inhibitor (NEB, catalog number: #M0314)
dNTP mix (Thermo Scientific, catalog number: R0192)
Random hexamer (Invitrogen, catalog number: N8080127)
Primers (IDT, origin: USA)
SYBR Fast qPCR Master Mix (KAPA Biosystem, catalog number: KK4602)
Solutions
Complete media for GM12878 cell line (see Recipes)
Hypotonic buffer (see Recipes)
RNA precipitation solution (RPS) (see Recipes)
Recipes
Complete media for GM12878 cell line
Reagent Stock concentration Final concentration Volume (mL) RPMI media 395 FBS 100% 20% 100 Penicillin-Streptomycin 10,000 U/mL 100 U/mL 5 Total 500 Hypotonic buffer
Reagent Stock concentration Final concentration Volume (μL) Tris (pH 7.5) 100 mM 10 mM 500 KCl 100 mM 10 mM 500 MgCl2 150 mM 1.5 mM 50 DTT 100 mM 0.5 mM 25 NP-40 10% 0.075% 37.5 Ribonucleoside vanadyl complexes 200 mM 2 mM 50 H2O 3837.5 Total 5,000 RNA precipitation solution (RPS)
Reagent Stock concentration Final concentration Volume (mL) Sodium acetate (pH 5.5) 3 M 150 mM 0.5 Ethanol 99.5% 9.5 Total 10 Final concentration of each component is 0.15 M sodium acetate (pH 5.5) in ethanol.
Keep at -20 °C for storage.
Equipment
Refrigerated tabletop centrifuge (15 or 50 mL conical tube adaptors) (Thermo Scientific, model: Multifuge X1R)
Refrigerated benchtop centrifuge (1.5 mL tube rotor) (Eppendorf, model: 5424R)
Light microscope (Olympus, origin: USA)
Vortexer (Biosan, model: V-1 plus)
Micropipettors (Gilson, origin: USA)
NanoDrop 2000c UV-Vis Spectrophotometer (Thermo Scientific, origin: USA)
Applied Biosystems Veriti Dx 96-Well Thermal Cycler PCR Thermocycler (Applied Biosystem, origin: USA)
Bio-Rad Connect Real-Time PCR System (Bio-Rad, origin: USA)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Tan, K. E., Ng, W. L., Ea, C. K. and Lim, Y. Y. (2023). Detection of Cytoplasmic and Nuclear Circular RNA via RT-qPCR. Bio-protocol 13(17): e4798. DOI: 10.21769/BioProtoc.4798.
分类
分子生物学 > RNA > qRT-PCR
细胞生物学 > 细胞器分离 > 分级分离
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link