- EN - English
- CN - 中文
Fast and Sustainable Thermo-osmotic DNA Extraction Protocol for Trans-spectrum Contingency and Field Use
用于跨谱应急和现场使用的快速、可持续的热渗透 DNA 提取方案
发布: 2023年09月05日第13卷第17期 DOI: 10.21769/BioProtoc.4796 浏览次数: 1724
评审: Shyam SolankiOlga SinAnonymous reviewer(s)

相关实验方案

优化高分子量 DNA 提取方法以用于 Magnaporthaceae 及其他禾本科根部真菌的长读长全基因组测序
Michelle J. Grey [...] Mark McMullan
2025年03月20日 3252 阅读
Abstract
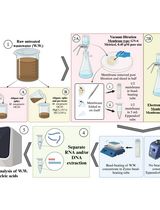
In the field of molecular genetics, DNA extraction protocols and kits are sample-specific and proprietary, preventing lateral distribution among similar facilities from different sectors to alleviate supply shortages during a crisis. Expanding upon previous fast extraction protocols such as alkaline- and detergent-based ones, the use of boiling-hot water to rupture cells, virions, and nuclei, as proposed during the COVID-19 pandemic, might alleviate shortages and costs. Different soft, relatively abundant (highly enriched), and uncomplicated (genomically homogenous and with few inhibitors) biosamples are collected in 1.5 mL tubes, mixed with boiling-hot water, and stirred vigorously, so as to have membranes lysed and proteins deactivated; mechanical disruption may be used as well if necessary. Incubation in boiling water bath for 20–30 min follows. Depending on sample type and quantity, which affects the total extraction volume, 2–5 μL are pipetted off for direct PCR and the same volume for two decimal serial dilutions. The latter are intended to optimize the crude extract to a workable DNA/inhibitor concentration balance for direct PCR. Uncomplicated, highly enriched samples such as mycelial growth in fruits and human swab samples can be processed, contrary to complicated samples such as blood and physically unyielding samples such as plant tissue. The extract can be used for immediate PCR in both benchtop and portable thermocyclers, thus allowing nucleic acid amplification tests (NAAT) being performed in resource-limited settings with low cost and waste footprint or during prolonged crises, where supply chain failures may occur.
Key features
• DNA extraction from different sample types using only boiling water and occasional mechanical assistance.
• Crude extract serially diluted twice, 10- and 100-fold, to bypass purification and quantification steps.
• Direct PCR for 2–10 μL of crude lysate and dilutions (conditional to sample type and quantity) to enhance probability of workable DNA-inhibitors’ concentrations.
• Lowers the cost and curtails the overall footprint of testing to increase sustainability in field operations and in standard lab environments under supply chain derailment.
Keywords: Thermo-osmotic DNA extraction (嗜热DNA 提取)Background
A fast and massive DNA extraction is quintessential for nucleic acid amplification tests (NAAT), especially if performed under duress; the latter implies either field conditions in resource-limited settings (RLS), in routine or expedient setups, or in abnormal conditions with samples surging and/or supply pipelines malfunctioning due to corporate concerns or supply chain overextension/collapse. Thus, although the technology for DNA extraction systems achieves significant yields from complicated and/or miniscule samples, their use is expensive even if purchased in bulk to achieve economies of scale. Alkaline approaches were suggested for contingency (Goudoudaki et al., 2021) and field conditions (Priye et al., 2016), adaptable to the intended use (Goudoudaki et al., 2021). Still, specific chemicals were used, which may not be available in proper quantities once a health or ecological crisis prompts massive spatial dispersion and increased processing output of incoming samples, which may overload standard public health or agronomic facilities and infrastructures. Thus, ease of procurement and use by personnel moderately trained in this specific discipline but experienced in other sectors of biosciences may increase the processing rate of initial screening and offload work from dedicated infrastructures, restricting their involvement to tackle only unresolved or suspect samples. Last but not least, this almost in-situ processing approach may limit the need for dispatching samples to distant facilities, which is expensive, time consuming, creates biosafety and biosecurity risks, and may degrade the samples, thus de-valuing the actual diagnostic procedure.
By using only distilled water and heat, both easily available, there is no chemical waste and no need for special reagents, and thus for diverse stockpiles. The latter may be inexpensive and long-lead or long-expiration date, but their storage, even in normal conditions, takes space and there is a need for correct management of the stock. Also, the skills necessary are basic, found in everyone with the most elementary training in Biosciences. Thus, the overall footprint of the analysis, both before- and after-use, is kept minimal. This protocol is actually an expansion of a previous one, developed during the COVID-19 pandemic (Fomsgaard and Rosenstierne, 2020) as a result of kits supply restrictions (Benda et al., 2021), to include different extraction substrates and target genomes. It can be used in the field, given that consumables, heat, and electricity sources for portable thermocyclers (Kambouris et al., 2020) can be provided. Compared to the alkaline protocol (Goudoudaki et al., 2021), the thermo-osmotic protocol described herein requires even less reagents and produces no chemical waste, nor does it require stocking reagents that may expire and require specific storage. The protocol is named after the thermal deterioration of the membrane bonds allowing osmosis to perform the cell lysis and has been used already in one original publication (Goudoudaki et al., 2023).
Additionally, the present protocol may be used for improving the turnaround time and volume throughput in existing, indoor facilities. This comes at the expense of other performance metrics including sensitivity, which are markedly inferior compared to the ones achieved by dedicated DNA extraction protocols, even those of relatively low cost (Velegraki et al., 1999a and 1999b). Furthermore, facilities commandeered during crisis management may be among the beneficiaries. The method may be used for public health and environmental and agronomic emergencies (Kambouris et al., 2018), thus qualifying for compatibility with the One-Health framework and any other that may refer to genetic/genomic biomarkers that may be typed by basic PCR protocols, with or without follow ups such as, but not restricted to, restriction digestion (Velegraki et al., 1999a and 1999b; Arabatzis et al., 2004) and single-locus sequencing (Irinyi et al., 2015). The method, being plain and simple, shows moderate performance and should be used for highly enriched (containing high numbers of target genomes) and uncomplicated (without similar loads of pseudo-targets and inhibitors) samples.
Materials and reagents
The provisions below do not include expendables and instrumentation necessary for PCR and downstream procedures, such as gel electrophoresis.
Biological materials
Swab/scrap samples from human oral mucosa (tongue or inner cheek/gum)
Fungal mycelia grown on fruits and vegetables
Reagents
Commercially available distilled water for house use (i.e., steam ironing) from any convenience store and manufacturer, upon availability
Absolute ethanol, 99.8% denatured with IPA, MEK, and Bitrex pure (Panreac/Applichem, catalog number: 147194) or any other of comparable concentration (high purity is not essential)
Solutions
70% ethanol (see Recipes)
Recipes
70% ethanol
Reagent Final concentration Volume Ethanol (absolute) 70% 700 mL H2O 30% 300 mL Total n/a 1,000 mL
Laboratory supplies
Falcon-shaped plastic tube, 50 mL (KIMA Vacutest, catalog number: KIMA-17102-50)
Beaker, 1,000 mL (Hamed, catalog number: 303173)
Polypropylene floating rack, 8 positions (Flinn Scientific, catalog number: FB1671)
Pasteur pipettes, glass (Hamed, catalog number: 303107)
Rubber bulb for Pasteur pipettes (Isolab Laborgerate, catalog number: 084-03-001)
Pipette P20, 0–20 μL (Gilson PIPETMAN®, catalog number: F123600)
Pipette P200, 20–200 μL (Gilson PIPETMAN®, catalog number: F123601)
Pipette P1000, 200–1,000 mL (Gilson PIPETMAN®, catalog number: F123602)
Pipette tips, 0–200 μL (KIMA Vacutest, catalog number: KIMA-18260)
Pipette tips, 1,000 μL (KIMA Vacutest, catalog number: KIMA-18172)
Plastic microtubes, 1.5 mL (PierceTM, catalog number: 69715)
Plastic microtubes for PCR, 0.2 mL (Thermo ScientificTM, catalog number: AB0620)
Scalpel blades (no. 22 stainless disposable; FEATHER, lot 03067430) or box cutter (e.g., Q-Connect, catalog number: 233471)
Rack (50-position for 18 mm diameter microtubes) (Fruugo)
Hydrophilic cotton (e.g., pharmacy package)
Plastic spoons (e.g., any convenience store, 10 pc package), spoons (e.g., stainless steel 18/0 convenience store), glass slides (76 mm × 26 mm, Knittel Glass, catalog number: 303157), or wood tongue depressor (disposable, Heine, catalog number: 508424)
Plastic pellet pestles, blue polypropylene autoclavable (Sigma-Aldrich, catalog number: Z359947-100EA)
Cotton (any pharmacy/convenience store) or paper tissue/napkins (any convenience store)
Equipment
Camping gas stove (e.g., KEMPER, 12.3 cm × 12.3 cm × 21.7 cm, model: 06450208) or electric kitchen stove burner (e.g., Severin, model: 3519368)
Lighter (e.g., KEMPER, model: 10422)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Goudoudaki, S., Kambouris, M. E., Manoussopoulou, M., Patrinos, G. P., Velegraki, A. and Manoussopoulos, Y. (2023). Fast and Sustainable Thermo-osmotic DNA Extraction Protocol for Trans-spectrum Contingency and Field Use. Bio-protocol 13(17): e4796. DOI: 10.21769/BioProtoc.4796.
分类
分子生物学 > DNA > DNA 提取
微生物学 > 病原体检测 > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link