- EN - English
- CN - 中文
Fluorescent Biosensor Imaging of Nitrate in Arabidopsis thaliana
拟南芥中硝酸盐的荧光生物传感器成像
发布: 2023年08月20日第13卷第16期 DOI: 10.21769/BioProtoc.4743 浏览次数: 3631
评审: David PaulLip Nam LOHAnonymous reviewer(s)
Abstract
Nitrate (NO3–) is an essential element and nutrient for plants and animals. Despite extensive studies on the regulation of nitrate uptake and downstream responses in various cells, our knowledge of the distribution of nitrogen forms in different root cell types and their cellular compartments is still limited. Previous physiological models have relied on in vitro biochemistry and metabolite level analysis, which limits the ability to differentiate between cell types and compartments. Here, to address this, we report a nuclear-localized, genetically encoded fluorescent biosensor, which we named nlsNitraMeter3.0, for the quantitative visualization of nitrate concentration and distribution at the cellular level in Arabidopsis thaliana. This biosensor was specifically designed for nitrate measurements, not nitrite. Through genetic engineering to create and select sensors using yeast, Xenopus oocyte, and Arabidopsis expression systems, we developed a reversible and highly specific nitrate sensor. This method, combined with fluorescence imaging systems such as confocal microscopy, allows for the understanding and monitoring of nitrate transporter activity in plant root cells in a minimally invasive manner. Furthermore, this approach enables the functional analysis of nitrate transporters and the measurement of nitrate distribution in plants, providing a valuable tool for plant biology research. In summary, we provide a protocol for sensor development and a biosensor that can be used to monitor nitrate levels in plants.
Key features
• This protocol builds upon the concept of FRET biosensors for in vivo visualization of spatiotemporal nitrate levels at a cellular resolution.
• Nitrate levels can be quantified utilizing the biosensor in conjunction with either a plate reader or a fluorescence microscope.
Graphical overview
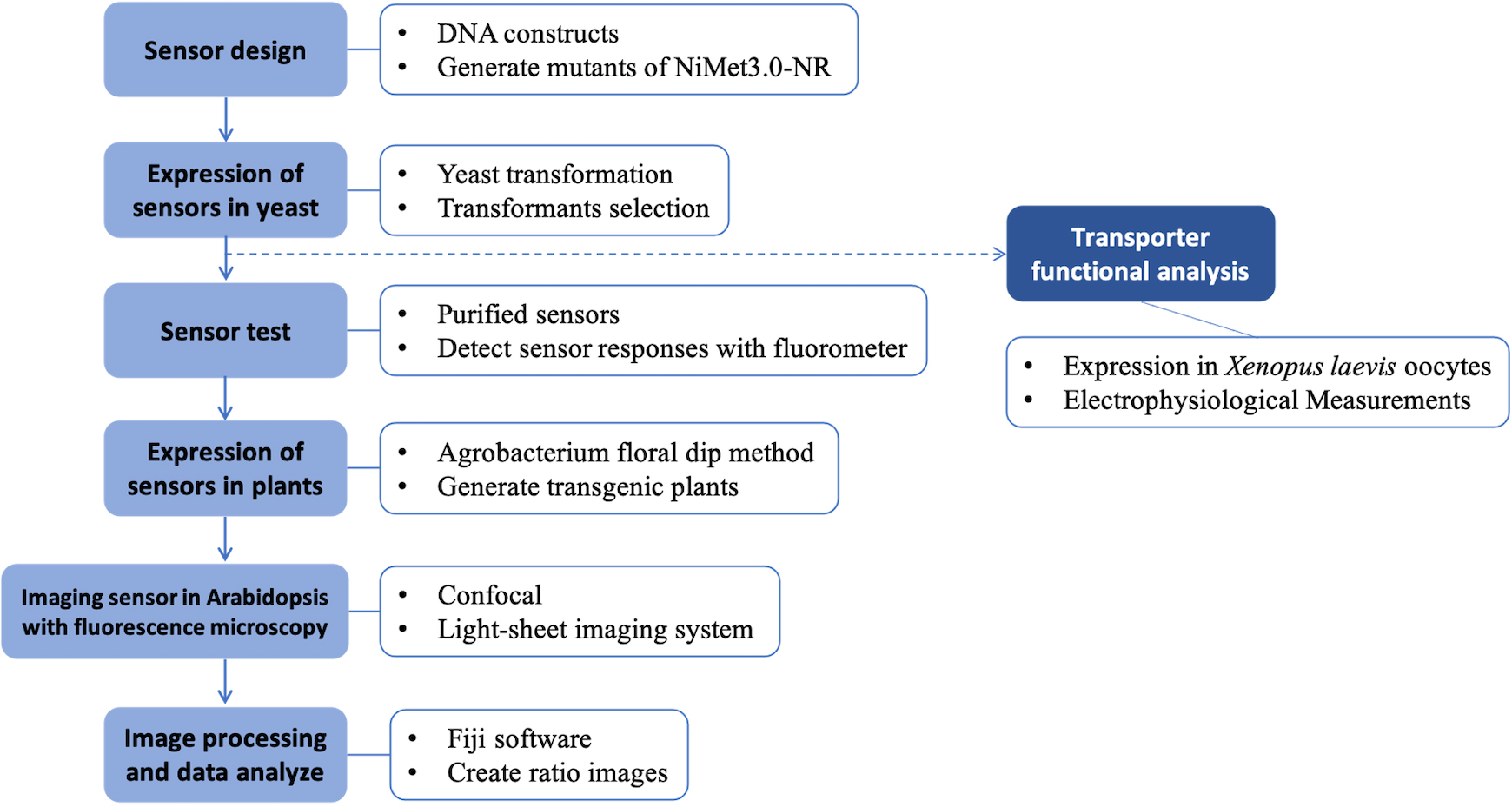
Background
Genetically encoded sensors have been developed over the past decade. The first fluorescence protein–based calcium sensor, Cameleon, was created and utilized to monitor calcium signaling processes in plant stomata (Allen et al., 2001). Since then, other sensors have been developed and applied in both plant and animal biology. One example is the glucose sensors FLIPglu600μΔ13V and FLIPsuc90μ∆1V, which were created by Chen et al. (2010 and 2012, respectively). These optical sensors have proven useful not only for monitoring analyte levels and fluxes but also for identifying elusive sucrose efflux transporters required for phloem loading. Overall, genetically encoded sensors have proven to be highly versatile tools with broad applicability in a range of research settings.
In our study, we developed a Förster resonance energy transfer (FRET)-based nitrate sensor, named nlsNitraMeter3.0, to successfully monitor the steady-state levels, accumulation, and dynamic conditions of nitrate distribution and content in Arabidopsis thaliana through fluorescence confocal microscopy. Using this sensor, we can directly visualize the spatial and temporal distribution of nitrate with high resolution at the cellular level. In addition, the sensor can be used to acquire images through fluorescence microscopy before and after treatment with different media. Through image analysis software (e.g., Fiji), the images can be quantified. Furthermore, the concept and operation of nlsNitraMeter3.0 can be applied to develop new sensors that identify or characterize other molecules in plants. For instance, we previously reported that NPF1.3 plays a role as a nitrate transporter by some functional in vitro analysis (e.g., Xenopus laevis oocytes) (Chen and Ho, 2022).
Here, we provide the principles of engineering, detection methods, and application of nlsNitraMeter3.0. By following the protocol reported herein it will also be possible to develop further sensors (refer to Supplementary information).
Materials and reagents
Biological materials
Yeast strain: protease-deficient yeast strain BJ5465 (MATa, ura3–52, trp1, leu2Δ1, his3Δ200, pep4::HIS3, prb1Δ1.6R, can1, GAL+) (ATCC, catalog number: 208289TM), which was obtained from the Yeast Genetic Stock Center (University of California, Berkeley, CA).
Sensors: NIT domain/NasR
The full-length open reading frame of NasR from Klebsiella oxytoca (Boudes et al., 2012) in the pDONR221 GATEWAY Entry vector was used as a sensory domain to create the nitrate sensors NiMet3.0 and nlsNiMet3.0. Note: Constructs were inserted using an Entry clone by Gateway LR reactions into the yeast expression vectors pDRFlip30.
The NiMet3.0 is fused to the full-length of NasR with pDRFlip30 vector and sandwiched between an N-terminal Aphrodite t9 (AFPt9) variant (Deuschle et al., 2006), with nine amino acids truncated off the C terminus, and a C-terminal monomeric Cerulean (mCer) (Rizzo et al., 2006).
Note: The pDRFlip30 vector was modified from pDRFlip39 vector (Addgene, catalog number: 65517).
Agrobacterium strain: Agrobacterium tumefaciens strain GV3101 was used here to obtain high transformation rates and high levels of expression, typically leading to high copy insertion into the genome (Koncz and Schell, 1986).
Arabidopsis thaliana: wild type Col-0, a nitrate transporter mutant [npf6.3/NTR1;1/chl1-5 (Leran et al., 2014)], and a nitrate reductase mutant [nia1nia2 (Desikan et al., 2002)] were used.
Reagents
Potassium nitrate (KNO3) (Sigma-Aldrich, catalog number: P8394)
Potassium chlorate (KClO3) (STREM, catalog number: 93-1913)
Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P5405)
Potassium nitrite (KNO2) (ARO, catalog number: 42306)
Potassium sulfate (K2SO4) (SHOWA, catalog number: 1648-4150-000-23)
Potassium sulfite (K2SO3) (ACROS, catalog number: 44021)
Potassium selenite (K2SeO3) (STREM, catalog number: 931971-000000-18)
Potassium molybdenum oxide, anhydrous (K2MoO4) (Alfa, catalog number: 22898-0000000-17)
Ammonium chloride (NH4Cl) (Merck, catalog number: 1.01145-0500)
Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M9272)
Gly-Gly (Sigma-Aldrich, catalog number: G1002)
YNB, yeast nitrogen base w/o amino acids w/o ammonium sulfate (BD, Difco, catalog number: 233520)
DO supplement-Ura (Takara Bio Company, Clontech, catalog number: 630416)
D-(+)-Glucose monohydrate (Fluka Analytical, catalog number: 49159)
Sucrose (Merck, catalog number: 1.07687.1000)
Agar (BD BactoTM, catalog number: DIF214530)
Agar (PhytoagarTM, catalog number: 40100072)
MES hydrate (Sigma-Aldrich, catalog number: M2933)
MOPS (Sigma-Aldrich, catalog number: M3183)
Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S5881)
1,4-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: DTT-RO)
Carrier DNA [UltraPureTM salmon sperm DNA solution (Thermo Fisher Scientific, InvitrogenTM, catalog number: 115632-011)]
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E6758)
Polyethylene glycol 4000 (PEG4000) (Fluka Analytical, catalog number: 81240)
Lithium acetate dihydrate (LiOAc) (Sigma-Aldrich, catalog number: L4158)
Tris hydrochloride ultrapure bioreagent (Tris-Cl) (J.T. Baker, catalog number: 4103-02)
Sodium chloride (NaCl) (Merck, catalog number: 1.06404.1000)
Sodium hydroxide (NaOH) (Merck, catalog number: 1.06498.1000)
Peptone (BD BactoTM, catalog number: 211677)
Yeast extract (BD BactoTM, catalog number: 212750)
MS modified basal salt mixture without nitrogen (MS) (PhytoTech Labs, catalog number: M531)
MilliQ or distilled water
Spectinomycin
Kanamycin
Solutions
40% (w/v) glucose solution (sterile and filtrated)
MOPS buffer
MES buffer
Yeast extract peptone dextrose (YPD) medium (see Recipe 1)
Solid yeast nitrogen base (see Recipe 2)
PLATE mixture (the acronym of PEG, lithium acetate, Tris, and EDTA) (see Recipe 3)
Wash buffer (see Recipe 4)
Resuspension buffer (see Recipe 5)
Substrate addition (see Recipe 6)
Plant growth solid base (see Recipe 7)
Recipes
Yeast extract peptone dextrose (YPD) mediuma, c
Reagent Final concentration Amount Peptone 2.0% (w/v) 10 g Yeast extract 1.0% (w/v) 5 g Agar 2.4% (w/v) 12 g 40% sterile filtrated glucoseb 2% (w/v) 25 mL H2O n/a n/a Total n/a 500 mL Autoclave, 121 °C, 15 psi, 15 min
For liquid medium, when hand-warm, add glucose from 40% sterile filtrated stock to a final concentration of 2% under a sterile hood (e.g., biosafety cabinet).
For solid medium, add 20 g/L agar before autoclaving. Add sterile filtrated glucose from 40% stock to a final concentration of 2% when the medium is hand-warm before pouring plates.
Solid yeast nitrogen base (-ura DropOut medium)a, c
Reagent Final concentration Amount Yeast nitrogen base w/o amino acids w/o ammonium sulfate 1.7 g/L 1.7 g DO supplement-Urad 0.77 g/L 0.77 g 40% sterile filtrated glucoseb 2% (w/v) 50 mL H2O n/a to 1,000 mL Total n/a 1,000 mL Autoclave, 121 °C, 15 psi, 15 min
For liquid medium, when hand-warm, add glucose from 40% sterile filtrated stock to a final concentration of 2% under a sterile hood (e.g., biosafety cabinet).
For solid medium, add 20 g/L agar before autoclaving. Add sterile filtrated glucose from 40% stock to a final concentration of 2% when the medium is hand-warm before pouring plates.
Adjust the pH of the -ura DropOut medium to pH 5.8 with NaOH before addition of agar and autoclaving.
PLATE mixture for yeast transformation
Reagent Amount 45% PEG4000 90 mL 1 M LiOAc 10 mL 1M Tris-Cl (pH7.5) 1 mL 0.5M EDTA 0.2 mL Total 100 mL Wash buffer
Reagent Final concentration Amount MES 50 mM 9.76 g Total n/a 1,000 mL Adjust the pH of the MES buffer to pH 5.5 with NaOH and autoclave.
Autoclave, 121 °C, 15 psi, 15 min.
Resuspension buffer
Reagent Final concentration Amount MES buffer (Recipe 4) 50 mM 250 mL Agar 0.05% 0.125 g Total n/a 250 mL Wait until the medium cools to room temperature (RT) to delay sedimentation of the cells during the measurement.
Plant growth solid basea–d
Reagent Final concentration Amount MS salts without nitrogen 1/2 strength 0.78 g 20% sterile filtrated sucrose 0.5 % (w/v) 25 mL Total n/a 1,000 mL Adjust the pH to pH 5.5 with KOH before addition of agar and autoclaving.
Autoclave, 121 °C, 15 psi, 15 min
For liquid medium, when hand-warm, add sucrose from 20% sterile filtrated stock to a final concentration of 0.5% under a sterile hood (e.g., biosafety cabinet).
For solid medium, add 12 g/L PhytoagarTM before autoclaving. Add sterile filtrated glucose from 20% stock to a final concentration of 0.5% when the medium is hand-warm before pouring plates.
Substrate addition
Reagent Final concentration Amount KNO3 or other substrates (Reagent 1–11) Depending on the experimental design - MES buffer 50 mM - Total n/a 50 mL Depending on the concentration of the nitrate needed, use the nitrate stock solution and dilute it with MES buffer or MOPS buffer.
Laboratory supplies
96-well microplates (flat bottom clear or black) (Greiner Bio-One, catalog numbers: 650101 and 650209)
Note: Black plates have a lower background.
Multichannel (12) pipette (for 100 μL) (e.g., Sartorius, catalog number: 725240)
50 mL sterile plastic tubes (Falcon®)
Petri dishes (diameter: 9 mm; height: 15 mm; sterile) (Alpha Plus, catalog number: BL6905)
Glass beads (3 mm) (BasicLife, catalog number: CEO-1169)
Vacuum-driven filter system (250 mL, upper cup, 0.22 μm PES) (AGC, catalog number: AGC-VC-PES22-250-1CS)
Equipment
Monochromator-based spectrofluorimeter for 96-well plates [Safire or Infinite® M1000 (Tecan Trading)]
Cell density meter (Amersham, model: Ultrospec 10)
Orbital shaker, with temperature and velocity control (Eppendorf, New Brunswick Scientific, model: Innova 44)
Incubator for 28–30 °C incubation of yeast cells (YIHDER, model: LM-570RD)
Centrifuge with swinging rotor for 50 mL tubes (Eppendorf, model: 5810R)
Inverted confocal plus super resolution microscope (Zeiss, LSM 780 + ELYRA): high sensitivity confocal microscope is equipped with GaAsP spectrum detector, and the super resolution microscope is a Structure Illumination Microscopy.
The laboratory-established light-sheet system was made in cooperation with Microlambda Pte Ltd (Singapore)
Growth chamber
Software and datasets
MetaMorph software (Downingtown, PA)
Fiji (http://fiji.sc/)
GraphPad Prism version 9.0.0 for Mac (www.graphpad.com)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chen, Y. N. and Ho, C. H. (2023). Fluorescent Biosensor Imaging of Nitrate in Arabidopsis thaliana. Bio-protocol 13(16): e4743. DOI: 10.21769/BioProtoc.4743.
- Chen, Y. N., Cartwright, H. N. and Ho, C. H. (2022). In vivo visualization of nitrate dynamics using a genetically encoded fluorescent biosensor. Sci Adv 8(42): eabq4915.
分类
植物科学 > 植物生物化学 > 代谢物
生物化学 > 蛋白质 > 荧光
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link