- EN - English
- CN - 中文
In situ Quantification of Cytosine Modification Levels in Heterochromatic Domains of Cultured Mammalian Cells
培养哺乳动物细胞异染色质中胞嘧啶修饰水平的原位定量分析
(*contributed equally to this work) 发布: 2023年07月20日第13卷第14期 DOI: 10.21769/BioProtoc.4716 浏览次数: 1711
评审: Gal HaimovichVeena PadmanabanAnonymous reviewer(s)

相关实验方案

基于Fiji ImageJ的全自动化流程开发:批量分析共聚焦图像数据并量化蛋白共定位的Manders系数
Vikram Aditya [...] Wei Yue
2025年04月05日 2841 阅读
Abstract
Epigenetic modifications of DNA, and especially cytosine, play a crucial role in regulating basic cellular processes and thereby the overall cellular metabolism. Their levels change during organismic and cellular development, but especially also in pathogenic aberrations such as cancer. Levels of respective modifications are often addressed in bulk by specialized mass spectrometry techniques or by employing dedicated ChIP-seq protocols, with the latter giving information about the sequence context of the modification. However, to address modification levels on a single cell basis, high- or low-content microscopy techniques remain the preferred methodology. The protocol presented here describes a straightforward method to detect and quantify different DNA modifications in human cell lines, which can also be adapted to other cultured mammalian cell types. To this end, cells are immunostained against two different cytosine modifications in combination with DNA counterstaining. Image acquisition takes place on a confocal microscopy system. A semi-automated analysis pipeline helps to gather data in a fast and reliable fashion. The protocol is comparatively simple, fast, and cost effective. By employing methodologies that are often well established in most molecular biology laboratories, many researchers are able to apply the described protocol straight away in-house.
Keywords: DNA methylation (DNA甲基化)Background
Methylation of the fifth carbon atom of cytosine (5′-methylcytosine; 5mC) is found in the DNA of most eukaryotes and in all human cell types. It is one of the most abundant and conserved epigenetic modifications and regulates gene expression by silencing promoter regions and facilitating the compaction of heterochromatin (Lande-Diner et al., 2007; Smith and Meissner, 2013). Mammalian DNA methylation occurs predominantly in the context of symmetric CpG dinucleotides. Consequently, of the 28 million CpG sites in the human genome, between 60% and 80% are methylated, which corresponds to approximately 5% of all cytosines (Smith and Meissner, 2013). However, up to 200 base-pair long stretches of these dinucleotides, so-called CpG-islands, are often found in the context of promoters and transcription start sites, where they are sparsely methylated (Bird, 1986; Smith and Meissner, 2013).
Moreover, DNA methylation contributes to the integrity of heterochromatin by serving as a binding platform for various proteins that can recognize and bind 5mC, thereby shaping the heterochromatin landscape (Ludwig et al., 2016). On the other hand, the members of the Tet (Ten-eleven-translocation)-protein family, Tet1, Tet2, and Tet3, were found to oxidize 5mC to 5′-hydroxymethylcytosine (5hmC), 5′-formylcytosine (5fC), and 5′-carboxylcytosine (5caC). All oxidative derivatives of 5mC were found to act as intermediates in the active removal of DNA methylation and are themselves removed via different glycosylases (Ludwig et al., 2016). Tet proteins share the same catalytic activity (Bauer et al., 2015) but different physiological roles (Santiago et al., 2014). An example is Tet1 and its short isoform, which exhibit distinct nuclear localization during DNA replication. This results in aberrant cytosine modification levels in heterochromatic regions of mouse and human cells and LINE 1 activation (Arroyo et al., 2022). While in murine cells heterochromatin rich domains are found on most chromosomes, in human cells long stretches of pericentric heterochromatin are mostly found on chromosomes 1, 9, and 16 (Meneveri et al., 1993; Bizhanova and Kaufman, 2021).
Changes in cytosine modifications and, in particular, hypomethylation, are a hallmark of many cancers (Vidal et al., 2017). A reproducible method to measure changes in cytosine modification levels in euchromatic vs. heterochromatic regions is highly desirable in this context. For this purpose, microscopy techniques are more revealing than mass spectrometry or ChIP-seq methods, since direct images from single cells provide information about the abundance, subnuclear distribution, and cell cycle variation of different epigenetic marks. Therefore, we developed a pipeline to address cytosine modification levels and applied it to MCF7 human breast adenocarcinoma cells, expressing higher levels of TET1 proteins, and MCF10a non-transformed human breast epithelial cells (Figure 1). Our method, performed in Arroyo et al. (2022), includes immunodetection of cytosine modifications with antibodies against 5mC and either 5hmC, 5fC, or 5caC, followed by confocal microscopy imaging and image analysis. The implemented image analysis includes nucleus and heterochromatin and (euchromatic) nucleoplasm segmentation, achieved by creating different masks based on the DNA counterstaining (DAPI) signal. Heterochromatin segmentation using DAPI as a proxy for differences in chromatin compaction has been extensively used in quantitative analyses of the nuclear landscape (Schmid et al., 2017; Pradhan and Cardoso, 2023). The following protocol can be applied to a variety of experimental models and cell lines in order to study and quantify differences in cytosine modification patterns in different cellular backgrounds.
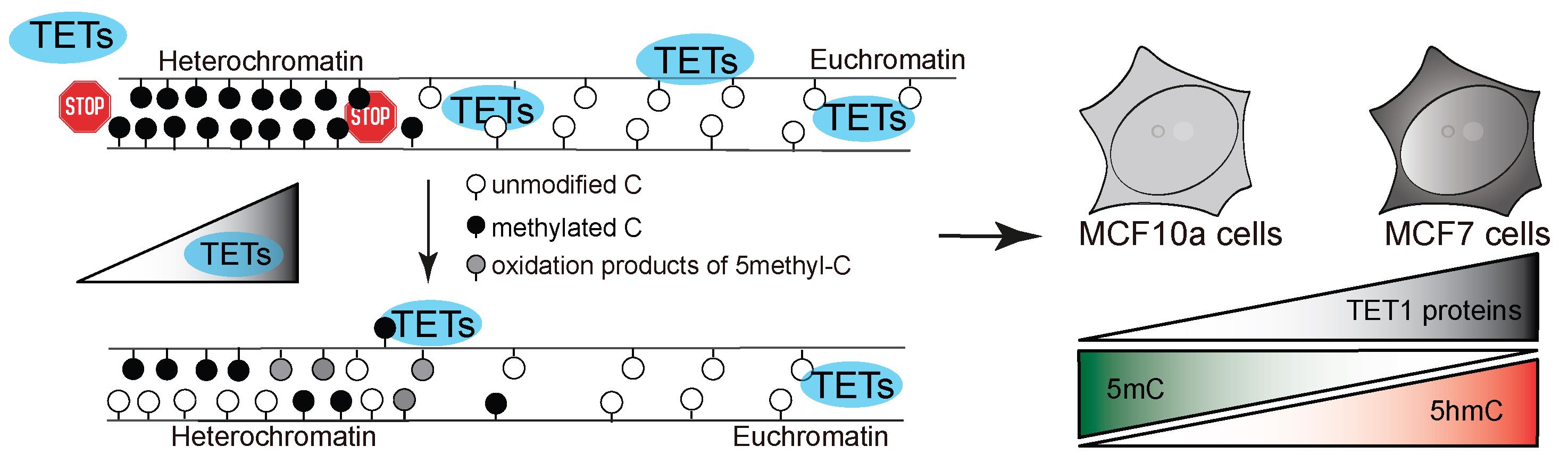
Figure 1. Model of study. TET protein catalytic activity in heterochromatin regions results in aberrant oxidation of 5-methylcytosine in human cells, depending on the level of TET proteins.
Materials and reagents
P100 dishes (Merck KGaA, catalog number: CLS430293)
6-well dishes (Merck KGaA, catalog number: CC302)
Glass coverslips (Carl Roth, catalog number: P232.1)
Microscopy slides (Thermo Fisher Scientific, catalog number: ABAA000080)
Petri dish (Carl Roth, catalog number: EL50.1)
Gel-blotting paper (Carl Roth, catalog number: A126.1)
Aluminum foil (Carl Roth, catalog number: 1770.1)
Parafilm (Merck KGaA, catalog number: P7793)
MCF7 cells (ATCC, catalog number: HTB-22) (Soule et al., 1973)
MCF10a cells (ATCC, catalog number: CRL-10317) (Soule et al., 1990)
DMEM (Merck KGaA, catalog number: D6429), store at 4 °C
DMEM/F12 (Merck KGaA, catalog number: D6421), store at 4 °C
Fetal bovine serum (FBS) (Gibco, catalog number: A5256701), aliquot and store at -20 °C
Horse serum (Merck KGaA, catalog number: H1138), aliquot and store at -20 °C
Pen/Strep (Merck KGaA, catalog number: P4333), store at 4 °C
L-Glutamine (Merck KGaA, catalog number: G7513), store at 4 °C
hEGF (Merck KGaA, catalog number: E9644)
Hydrocortisone (Merck KGaA, catalog number: H0888)
Cholera toxin (Merck KGaA, catalog number: C8052)
Insulin human (Merck KGaA, catalog number: I2643)
PBS (VWR International, catalog number: 0780)
Trypsin (Merck KGaA, catalog number: T4049), store at 4 °C
Gelatine (Sigma-Aldrich, catalog number: G2500)
Formaldehyde (Merck KGaA, catalog number: F8775)
EDTA (Merck KGaA, catalog number: E5134)
MgCl2·6H2O (Merck KGaA, catalog number: 1374248)
CaCl2 (Merck KGaA, catalog number: C1016)
Trizma® hydrochloride (Merck KGaA, catalog number: T2319)
Tween 20 (Merck KGaA, catalog number: P9416)
Triton X-100 (Merck KGaA, catalog number: X100)
Methanol (Merck KGaA, catalog number: 32213)
RNase A (Merck KGaA, catalog number: 10109169001)
BSA (Merck KGaA, catalog number: 10735094001)
DNase I (Merck KGaA, catalog number: D5025)
Monoclonal mouse α-5mC (CF-MAB - Helmholtz Munich, 32E2) (Weichmann et al., 2020)
Polyclonal rabbit α-5-hmC (Active Motif, catalog number: 39769)
Polyclonal rabbit α-5-fC (Active Motif, catalog number: 61223)
Polyclonal rabbit α-5-caC (Active Motif, catalog number: 61225)
Goat α-mouse IgG (H+L) Alexa Fluor 488-conjugated (Thermo Fisher Scientific, catalog number: A32723) (suggested antibody, which worked well in this protocol)
Goat α-rabbit IgG (H+L) Alexa Fluor 594-conjugated (Thermo Fisher Scientific, catalog number: A32740) (suggested antibody, which worked well in this protocol)
DAPI, 4’,6-diamidino-2-phenylindole (Merck KGaA, catalog number: D9542)
Mowiol® 4-88 (Merck KGaA, catalog number: 81381)
Wet chamber for staining (Cardoso and Leonhardt, 1996)
Solutions
0.1% gelatine (see Recipes)
3.7% formaldehyde (see Recipes)
88% methanol (see Recipes)
0.5% Triton X-100 in PBS (see Recipes)
0.01% PBS-T (see Recipes)
0.01% PBS-TE (see Recipes)
4%, 2%, 1% BSA in PBS (see Recipes)
100 µg/mL RNase in 1× PBS (see Recipes)
2× DNase I Buffer (see Recipes)
Recipes
0.1% gelatine
Reagent Final concentration Amount Gelatine 0.1% 0.5 g 1× PBS 1× 500 mL Total n/a 500 mL Sterilize by autoclaving at 121 °C for 30 min. 3.7% formaldehyde
Reagent Final concentration Amount 36.5%–38% formaldehyde solution 3.7 % 2.5 mL 1× PBS 1× 22.5 mL Total n/a 25 mL Prepare fresh and use within one week. 88% methanol
Reagent Final concentration Amount Methanol 88% 88 mL ddH2O 12% 12 mL Total n/a 100 mL Store at -20 °C 0.5% Triton X-100 in PBS
Reagent Final concentration Amount 1× PBS 1× 99.5 mL Triton X-100 0.5% 0.5 mL Total n/a 100 mL Prepare at least two days prior to use and store at room temperature. 0.01% PBS-T
Reagent Final concentration Amount 1× PBS 1× 249.975 mL Tween 20 0.01% 0.025 mL Total n/a 250 mL Store at room temperature. 0.01% PBS-TE
Reagent Final concentration Amount 1× PBS 1× 249.75 mL Tween 20 0.01% 0.025 mL EDTA 100 mM Total n/a 250 mL Store at room temperature 4% BSA in PBS
Reagent Final concentration Amount BSA 4% 0.6 g 1× PBS 1× 15 mL Total n/a 15 mL Dissolve BSA completely, prepare 1.5 mL aliquots, and store at -20 °C. If 1% or 2% BSA are needed, prepare appropriate dilutions with 1× PBS. 100 µg/mL RNase in 1× PBS
Reagent Final concentration Amount RNase A - 0.025 g ddH2O - 250 µL Total 100 mg/mL 250 µL Dissolve RNase A powder completely in ddH2O and store in 25 µL aliquots at -20 °C.
Before use, prepare a 1:1,000 dilution with 1× PBS to yield a concentration of 100 µg/mL.2× DNase I Buffer
Reagent Final concentration Amount Trizma® hydrochloride pH 7.5 20 mM 2 mL MgCl2 5 mM 0.102 g CaCl2 1 mM 0.011 g ddH2O - 98 mL Total n/a 100 mL Prepare 2 mL aliquots and store at -20 °C.
Equipment
Laminar flow cabinet (neoLab Migge GmbH, BDK 1200-SK, catalog number: 83 99 51200)
Cell culture CO2 incubator (Binder, catalog number: C150-230V)
Water bath (GFL Technology, catalog number: 10865892E)
Hybridization oven (Appligene, catalog number: 22012105); alternatively, use 37 °C incubator
Confocal microscope (Perkin Elmer Spinning Disk/Nikon Ti-E, PerkinElmer Life Sciences) equipped with high magnification objective (oil immersion 100× Plan-Apochromat, NA 1.49)
Software
Image acquisition software [in our case, Volocity 6.3 (Perkin Elmer, USA) was used] “Fiji Is Just ImageJ” Fiji (NIH/http://Fiji.sc/Fiji) or ImageJ (NIH/https://imagej.nih.gov/ij/download.html) for image pre-processing and analysis
RStudio (https://rstudio.com/) for plotting and statistical analysis
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Arroyo, M., Cardoso, M. C. and Hastert, F. D. (2023). In situ Quantification of Cytosine Modification Levels in Heterochromatic Domains of Cultured Mammalian Cells. Bio-protocol 13(14): e4716. DOI: 10.21769/BioProtoc.4716.
分类
分子生物学 > DNA > DNA 修饰
细胞生物学 > 细胞成像 > 共聚焦显微镜
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link