- EN - English
- CN - 中文
Agrobacterium-mediated Genetic Transformation of Cotton and Regeneration via Somatic Embryogenesis
农杆菌介导的棉花遗传转化和通过体细胞胚胎发生的再生
发布: 2023年05月20日第13卷第10期 DOI: 10.21769/BioProtoc.4677 浏览次数: 4332
评审: Amey RedkarAnonymous reviewer(s)
Abstract
Cotton is a significant industrial crop, playing an essential role in the global economy that suffers several setbacks due to biotic and abiotic adversities. Despite such problems, biotechnological advances in cotton are limited because of genetic transformation and regeneration limitations. Here, we present a detailed protocol optimized based on previously published papers, along with our modifications. These involve changes in Agrobacterium concentration, co-cultivation time and temperature, hormones used for regeneration, media manipulation for embryogenic callus production, and efficient rescue of deformed embryos. Further, this protocol has been used in genetic studies on biotic and abiotic stress in cotton. This protocol assures a reproducible stable transgenic cotton development procedure via somatic embryogenesis that can be used by researchers worldwide.
Keywords: Cotton (棉花)Background
Genetic engineering has opened the doors to enhanced yield, disease resistance, and improved nutritional quality and quantity. Genetically modified (GM) crops ensure that the needs of the growing population are met worldwide. The altered attributes in GM crops are due to introduced transgenes, which is achieved by different tools as particle bombardment, chloroplast transformation, pollen tube pathway transformation, protoplast electroporation, Agrobacterium-mediated genetic transformation, and many more. Amongst these, Agrobacterium tumefaciens–mediated transformation is the preferred one due to its exceptional trait of transferring a DNA segment into the host cell via a specialized plasmid (Ziemienowicz, 2014). This technique has become favorable for scientists and industries because of its simple and easy approach, its ability to transfer a low copy number of the gene of interest, and its efficiency of transfer of a large DNA fragment into the host cell—all of this at a very low cost. Agrobacterium-mediated genetic transformation has been employed to transform many economically important crops using plant tissue culture, which helps to regenerate a large number of transgenic plants at a single time point (Somleva et al., 2002; Manickavasagam et al., 2004; Jones et al., 2005; Sun et al., 2006).
Transgenic cotton was one of the first genetically modified organisms to be globally accepted and commercially developed. Now and again, researchers have been trying to improve the quality of cotton crops, either by enhancing its fiber yield or by imparting pests resistance. Developing a transgenic cotton plant is tedious and time consuming because of its recalcitrant nature and high production of phenolics, which hinders the regeneration of the plant. Although Agrobacterium-mediated genetic transformation has enabled the development of stable transgenic cotton, various efforts have been made to improve the transformation process (Qandeel-E-Arsh et al., 2021). A successful transformation does not depend on a single parameter; instead, it heavily relies on multiple factors such as Agrobacterium strain, type and age of explants, duration of co-cultivation between Agrobacterium and the plant tissue, bacterial cell density during transformation, regeneration method for the plant, selection marker, and type of vector to use (Firoozabady et al., 1987; Satyavathi et al., 2002; Jadhav and Katageri, 2017). To develop transgenic cotton plants, scientists have used the hypocotyls, cotyledons, and shoot tips, as well as the embryogenic callus, pollen tube, embryo, and meristems as explants (Jin et al., 2005; Pathi and Tuteja, 2013; Wang et al., 2013). The use of meristems is very cumbersome and labor intensive and requires particle bombardment (with the drawback of high copy number). The use of the embryonic callus is a relatively good option, but these take several months to be obtained, and the subsequent process does not guarantee proper plantlet germination (McCabe and Martinell, 1993). Transformation of the shoot tip, the embryo, and the pollen tube is easier than others, but the potential for stable transgenic plants is low. Most of these techniques fall into the category of transient transformation, skipping the inserted gene in subsequent generations. Direct organogenesis has also been tried in order to facilitate regeneration, but when the transformation is involved, it becomes sluggish and dreary, and chimeric plants are often obtained. Many of these issues can be resolved by using hypocotyls or cotyledons as the explant, as the process involves somatic embryogenesis that produces somatic embryos, which are single cell–derived. Furthermore, this promotes high-end vegetative output, ensuring more transgenic plants.
Various regeneration protocols have been proposed by several researchers. They have fiddled with different concentrations and types of plant hormones to induce callus, manipulated growth media to induce embryogenesis in proliferating calli, and changed carbohydrate sources as well (Shoemaker et al., 1986; Finer, 1988; Hemphill et al., 1998; Aydin et al., 2004; Ikram-ul-Haq and Zafar, 2004; Kumar and Tuli, 2004). All these attempts have successfully regenerated cotton genotypes, but the development of stable transgenic lines requires an appropriate combination of cotton regeneration and transformation. Currently, the most commonly used approaches are the pollen tube pathway, embryo transformation, and meristem-mediated methods (Bajwa et al., 2015), but these do not guarantee stable gene transfer. Using these methods, elucidation of the applied hypothesis is possible, but only a few have been able to be applied in practical use for stable transgenic development.
Here, we propose a protocol for Agrobacterium-mediated genetic transformation and regeneration in cotton, resulting in transgenic production (Figure 1). This protocol is based on the approach described by Kumar and Tuli (2004), with various modifications that accelerate the achievement of the transgenic development. We have optimized the media for the Agrobacterium to be ready to infect the explants, the co-cultivation period, the appropriate quantity and type of phytohormones to be used, and the environment required for callus induction, its proliferation, and further embryogenesis, and we have reduced the abnormalities in the embryos produced. To summarize, this protocol is an approach towards obtaining a stable transgenic line, via transforming the explants with a gene of interest and regenerating them with early callus induction, improved embryogenesis, and culture requisites. We have used a 14 kb construct harboring a CamV35S promoter, the pectin methylesterase (PME) gene, the neomycin phosphotransferase II (nptII), and the nopaline synthase (Nos) terminator. The protocol has been used to develop several distinct transgenic lines and is currently being used to alter numerous unique genes in cotton for genome editing.
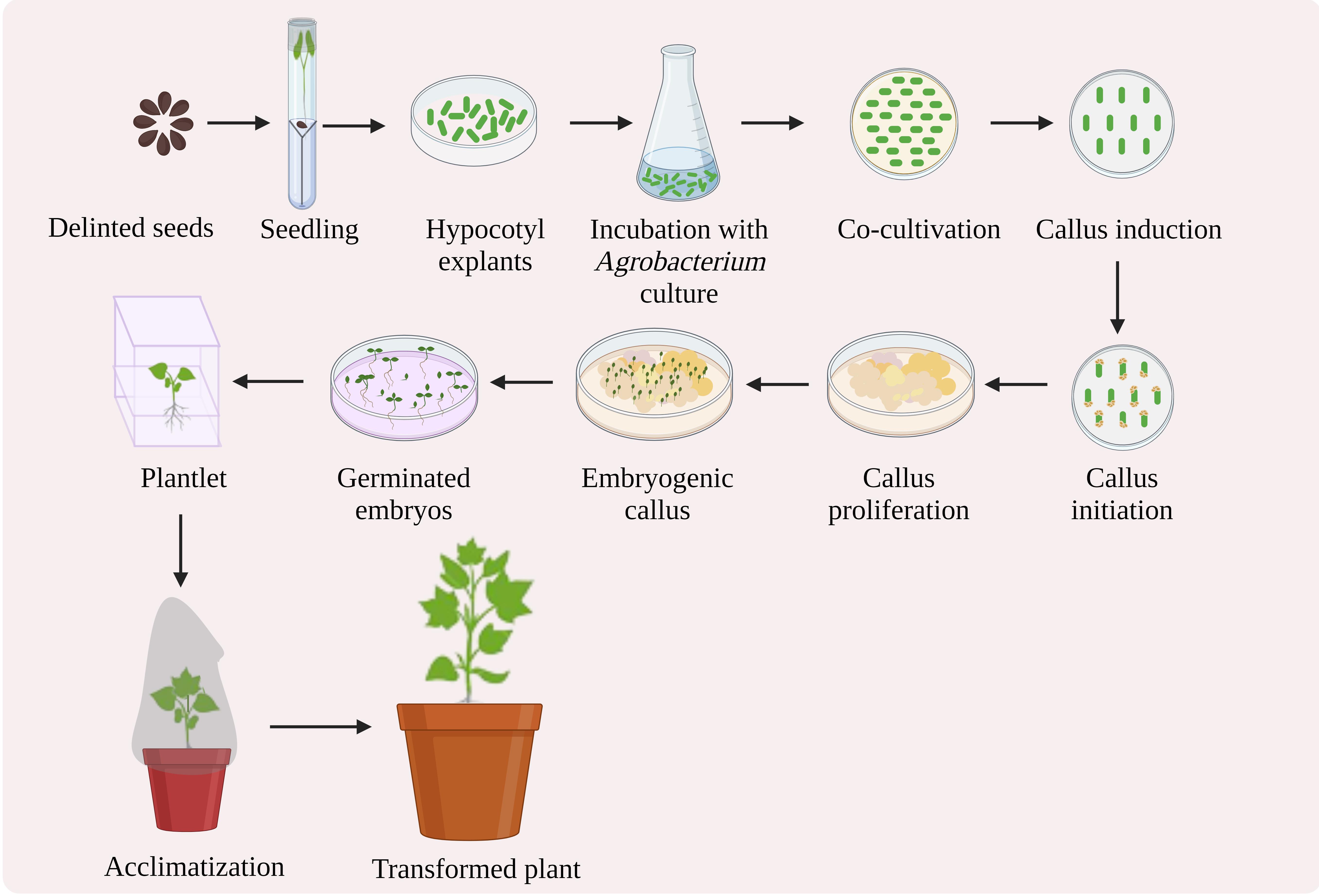
Figure 1. Schematic representation of Agrobacterium-mediated genetic transformation of cotton hypocotyl explants and regeneration (created with BioRender.com)
Materials and Reagents
Glassware
Glass rod
Conical flask, 250 mL (Borosil, catalog number: 4984021)
Glass bottle, 1,000 mL (Borosil, catalog number: 1501029)
Culture vials, 30 mL (Borosil, catalog number: 9910010)
Test tubes
Wide-mouth bottles, 100 mL
Plasticwares
Petri plates, 90 × 20 mm (Tarsons, catalog number: 960096)
Petri plates, 90 × 15 mm (Tarsons, catalog number: 960091)
Petri plates, 100 × 20 mm (Genetix, catalog number: 310100)
Parafilm tape (Tarsons, catalog number: 380020)
Stainless steel sieve
Filter paper (Whatman, catalog number: 1003-917)
Computer paper
Planton plant tissue culture container, 7.5 × 7.5 × 10 cm (Tarsons, catalog number: 020080)
Frames for planton boxes (Genetix, catalog number: 310074)
Spirit lamp
Plastic bags
Falcon tubes, 50 mL
Sterile syringe
Test tube caps (Tarsons, catalog number: 020070)
Plant earthen pots, 14”
Plant plastic pots, 5”
Sterile syringe filters, 0.22 μm
Electroporation cuvettes (gene pulser 0.4 cm gap) (Bio-Rad, catalog number: 1652086)
Toothpick
Scalpel
Forceps
Surgical blade
Biological materials
Matured, defuzzed, and delinted cotton seeds. Variety: Coker-312
Agrobacterium tumefaciens strain LBA4404
Binary vector incorporating gene of interest. Here, PME (pectin methylesterase gene) is cloned in pBI121 vector under the promoter CaMV35S, neomycin transferase II (nptII) as selectable marker, and Nos terminator.
Chemicals and reagents
Sodium bicarbonate (Himedia, catalog number: MB045), store at room temperature (RT)
Sulfuric acid (Himedia, catalog number: RM6224), store at RT
Mercuric chloride (HgCl2) (Sigma-Aldrich, catalog number: M1136), store at RT
Ethanol (Merck, catalog number: 1.00983), store at RT
Acetosyringone (Himedia, catalog number: PCT1301), store at 4 °C
Glucose (Sigma-Aldrich, catalog number: G7021), store at RT
Sucrose (Sigma-Aldrich, catalog number: S0389), store at RT
Phytagel (Sigma-Aldrich, catalog number: P8169), store at RT
Agar powder (Himedia, catalog number: MB053), store at RT
Myo-inositol (Sigma-Aldrich, catalog number: I7508), store at RT
Magnesium chloride (Sigma-Aldrich, catalog number: M4880), store at RT
Potassium chloride (Sigma-Aldrich, catalog number: P5405), store at RT
Calcium chloride (Sigma-Aldrich, catalog number: C5670), store at RT
MES buffer (Sigma-Aldrich, catalog number: M3671), store at RT
Murashige & Skoog basal salt (Sigma-Aldrich, catalog number: M5524), store at 4 °C
Potassium nitrate (Sigma-Aldrich, catalog number: P8291), store at RT
Magnesium sulfate (Sigma-Aldrich, catalog number: M2773), store at RT
Potassium phosphate monobasic (Sigma-Aldrich, catalog number: P5655), store at RT
Calcium nitrate tetrahydrate (Sigma-Aldrich, catalog number: C2786), store at RT
Ammonium phosphate monobasic (Sigma-Aldrich, catalog number: 216003), store at RT
Calcium chloride dehydrate (Sigma-Aldrich, catalog number: C7102), store at RT
Sodium molybdate dehydrate (Sigma-Aldrich, catalog number: M1651), store at RT
Boric acid (Sigma-Aldrich, catalog number: B7901), store at RT
Potassium iodide (Sigma-Aldrich, catalog number: 221945), store at RT
Manganese sulfate tetrahydrate (Sigma-Aldrich, catalog number: 1.02786), store at RT
Zinc sulfate (Sigma-Aldrich, catalog number: Z0251), store at RT
Copper sulfate pentahydrate (Sigma-Aldrich, catalog number: C8027), store at RT
Cobalt chloride hexahydrate (Sigma-Aldrich, catalog number: C8661), store at RT
Sodium EDTA (Sigma-Aldrich, catalog number: 03650), store at RT
Ferrous sulfate heptahydrate (Sigma-Aldrich, catalog number: F8633), store at RT
Nicotinic acid (Sigma-Aldrich, catalog number: N0761), store at RT
Pyridoxine HCl (Sigma-Aldrich, catalog number: P6280), store at RT
Thiamine HCl (Sigma-Aldrich, catalog number: T1270), store at RT
Kinetin (Sigma-Aldrich, catalog number: K3375), store at 4 °C
2,4-Dichlorophenoxyacetic acid (2,4-D) (Sigma-Aldrich, catalog number: D76724), store at 4 °C
6-Benzylaminopurine (BAP) (Sigma-Aldrich, catalog number: B3408), store at 4 °C
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418), store at RT
Methanol (Sigma-Aldrich, catalog number: 34860), store at RT
Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: 221465), store at RT
Yeast extract (Sigma-Aldrich, catalog number: Y1625), store at RT
Beef extract (Himedia, catalog number: RM002V), store at RT
Peptone (Sigma-Aldrich, catalog number: P5905), store at RT
Murashige & Skoog basal salt mixture (4.3 g/L) (Sigma-Aldrich; catalog number: M2909)
Antibiotics (see Recipes)
Kanamycin 50 mg/L (Himedia, catalog number: MD026), store at 4 °C
Streptomycin 250 mg/L (Himedia, catalog number: MD048), store at 4 °C
Rifampicin 50 mg/L (Himedia, catalog number: MD045), store at 4 °C
Augmentin 250 mg/L (Himedia, catalog number: PCT1115), store at 4 °C
Cefotaxime 250 mg/L (Himedia, catalog number: MD015), store at 4 °C
Phytohormones (see Recipes)
2,4-D (stock: 10 mg/mL; working: 0.5 mg/mL)
Kinetin (stock: 10 mg/mL; working: 0.2 mg/mL)
BAP (stock: 10 mg/mL; working: 0.2 mg/mL)
Acetosyringone (see Recipes)
YEB media (see Recipes)
Induction media (see Recipes)
Hoagland media (see Recipes)
Murashige & Skoog stock (see Recipes)
Culture media (see Recipes)
Equipment
Incubator/shaker (Kuhner, model: ISF-1-W)
Laminar flow hood
Autoclave (TOMY, model: ES-315)
Ultracentrifuge (Eppendorf, model: 5430R)
pH meter (Eutech, pH tutor)
Magnetic stirrer (Abdos, Swirl top)
Thermal cycler (Bio-Rad, C1000 thermal cycler)
-80 °C ultra-low temperature freezer (Thermo Scientific, Forma 89000 Series)
-20 °C low temperature freezer (Celfrost)
Weighing balance (Sartorius, SQP-F)
Electroporation machine (MicroPluserTM Bio-Rad)
Greenhouse
Culture room
Soil mix for plants
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Srivastava, A., Shukla, A. K., Srivastava, S., Dubey, R. S., Singh, P. K. and Verma, P. C. (2023). Agrobacterium-mediated Genetic Transformation of Cotton and Regeneration via Somatic Embryogenesis. Bio-protocol 13(10): e4677. DOI: 10.21769/BioProtoc.4677.
分类
植物科学 > 植物转化 > 农杆菌介导的转化方法
分子生物学 > DNA > 转化
生物科学 > 生物技术 > 微生物技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link