- EN - English
- CN - 中文
Visualizing the Cisternal Organization of Golgi Ministacks in HeLa Cells by Side-averaging
通过侧向平均方法来可视化HeLa细胞中高尔基Ministacks的池状组织
发布: 2023年04月20日第13卷第8期 DOI: 10.21769/BioProtoc.4658 浏览次数: 1201
评审: Ralph Thomas BoettcherSusanne ReinhardtTakashi Nishina
Abstract
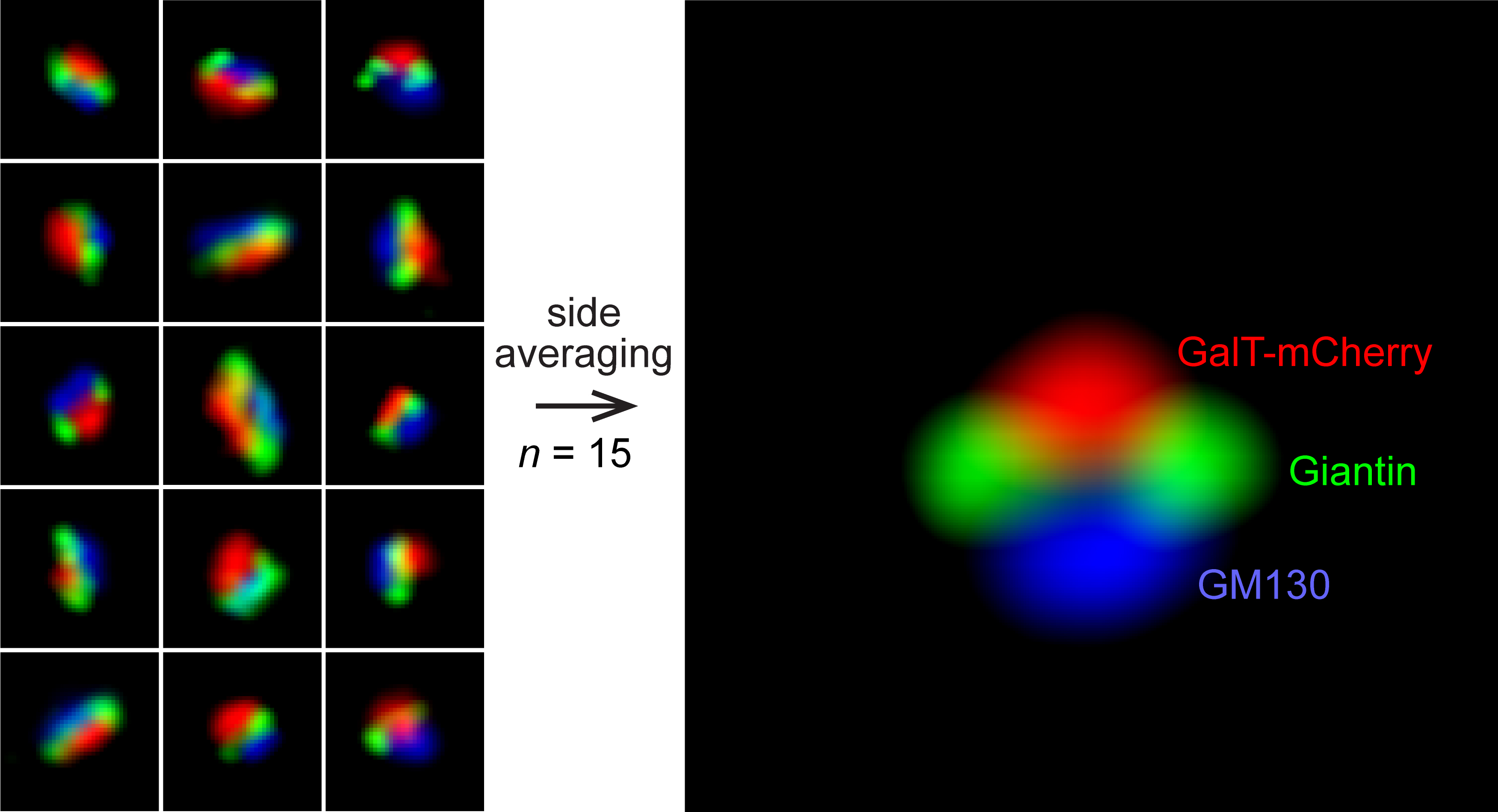
The mammalian Golgi complex consists of laterally connected Golgi stacks, each comprising close-packed and flattened membrane sacks called cisternae. However, the convoluted spatial organization of Golgi stacks and limited resolution of light microscopy prevent us from resolving the cisternal organization of the Golgi. Here, we describe our recently developed side-averaging approach coupled with Airyscan microscopy to visualize the cisternal organization of nocodazole-induced Golgi ministacks. First, the nocodazole treatment greatly simplifies the organization of Golgi stacks by spatially separating the crowded and amorphous Golgi complex into individual disk-shaped ministacks. The treatment also makes it possible to identify en face and side-views of Golgi ministacks. Next, after manually selecting the side-view Golgi ministack images, they are transformed and aligned. Finally, the resulting images are averaged to enhance the common structural features and suppress the morphological variations among individual Golgi ministacks. This protocol describes how to image and analyze the intra-Golgi localization of giantin, GalT-mCherry, GM130, and GFP-OSBP in HeLa cells by side-averaging.
Graphical overview
Background
The Golgi complex (Golgi) plays a central role in sorting and modifying secretory proteins and lipids (cargos) (Glick and Luini, 2011; Klumperman, 2011; Lu and Hong, 2014). In mammalian cells, the Golgi consists of a laterally linked network of Golgi stacks. As the structural unit of the Golgi, a Golgi stack comprises 4–7 flattened and densely packed membrane sacks called cisternae. The stacked cisternae of the Golgi can be further divided into three zones, namely the cis, medial, and trans-Golgi zones. At the trans-side of the Golgi stack, the trans-Golgi network localizes immediately outside the trans-Golgi zone and comprises tubular and vesicular membrane profiles. The secretory cargos synthesized in the ER reach the cis-Golgi and transit the medial before eventually exiting at the trans-side of the Golgi.
The structural unit of the Golgi, the Golgi stack, has an extremely diverse and complex morphology. It has an amorphous and twisted shape and many attached tubular and vesicular membrane profiles. The spatial resolution of the conventional light microscope is ~250 nm, which is insufficient to resolve the 200–400 nm thick Golgi stack. Therefore, under a fluorescence microscope, the mammalian Golgi appears as a perinuclear lump with almost unresolvable structural features. The difficulty in imaging the Golgi is also due to the dense and random orientation of the Golgi stacks. Hence, it is critical to perform multicolor imaging to identify different zones of the same Golgi stack. Although possessing a spatial resolution of 40–100 nm, super-resolution microscopic techniques such as stimulated emission depletion microscopy and single-molecule localization microscopy have difficulty resolving the Golgi's cisternal organizations, since it is very challenging for them to perform multicolor imaging (Schermelleh et al., 2019). In the past, conventional and super-resolution light microscopy have had only limited success in imaging the Golgi cisternae with primarily qualitative data, mostly revealing the relative distributions of no more than three Golgi proteins (Dejgaard et al., 2007; Bottanelli et al., 2016; Tie et al., 2016; Zhang et al., 2020; Hao et al., 2021).
Nocodazole treatment scatters the densely aggregated Golgi as disk-shaped individual structural units with relatively uniform morphological appearances. Extensive data have demonstrated that nocodazole-induced Golgi ministacks (hereafter Golgi ministacks) reliably represent the native Golgi stack (Rogalski et al., 1984; Van De Moortele et al., 1993; Cole et al., 1996; Trucco et al., 2004; Tie et al., 2018). Therefore, Golgi ministacks are a valid Golgi model. We developed a quantitative method called GLIM (Golgi localization by imaging the center of fluorescence mass) (Tie et al., 2016 and 2017). GLIM can achieve ~30 nm spatial resolution along the cis-trans Golgi axis. However, it only provides a numerical value denoting the axial localization but does not generate a pictorial representation of the cisternal distribution of Golgi proteins. By averaging en face views of Golgi ministacks, we later developed an en face averaging method to study the lateral organization of Golgi proteins (Tie et al., 2018). Recently, we have further advanced the averaging approach and introduced the side-averaging method to visualize the cisternal organization of Golgi ministacks directly (Tie et al., 2022). Averaging the Golgi ministack images can effectively enhance the common structural features and suppress the morphological variations or noises among individual Golgi ministacks. Side-averaging provides a systematic and quantitative imaging approach to reveal the axial and lateral cisternal distributions of Golgi proteins. Here, we present a detailed side-averaging protocol to image the intra-Golgi distribution of four Golgi proteins: giantin, GalT-mCherry, GM130, and GFP-OSBP.
Materials and Reagents
Cell culture materials and reagents
Φ 12 mm glass coverslip (no. 1.5) (Menzel, catalog number: CB00120RAC)
24-well cell culture plates (TC-treated and F-bottom) (Corning® Costar®, catalog number: 3524)
T25 cell culture flask (Nunc®, catalog number: 156367)
15 mL centrifuge tube (Greiner, catalog number: 188271)
0.45 µm syringe filter with Supor® membrane (25 mm, sterile) (Pall, catalog number: 4614)
Glass slide (3 × 1 inch) (Biomedia, catalog number: BMH.880103)
Kimwipe® (Kimtech, catalog number: 34155)
Dulbecco's modified Eagle's medium (DMEM) (Capricon, catalog number: DMEM-HPA-P50)
0.25% Trypsin-EDTA (Gibco, catalog number: 25200072)
Fetal bovine serum (FBS) (GE, Hyclone, catalog number: SV30160.03HI)
Opti-MEM (Invitrogen, catalog number: 31985070)
Lipofectamine 2000 (Invitrogen, catalog number: 116688-019)
Chemicals
Nocodazole (Merck, catalog number: 487928)
Dimethyl sulfoxide (DMSO) (Merck, catalog number: D8418-50ML)
Paraformaldehyde (Merck, catalog number: 1.04005.1000)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A9418)
Saponin (Sigma-Aldrich, catalog number: 47036)
Glycerol (Sigma-Aldrich, catalog number: G5516-1L)
Mowiol 4-88 (MW ~31,000) (Merck, catalog number: 81381-50G)
1,4-Diazabicyclo [2.2.2] octane (DABCO) (Merck, catalog number: D27802-25G)
Tris base (Promega, catalog number: H5135)
Sodium chloride (NaCl) (Merck, catalog number: 1.06404.5000)
Potassium chloride (KCl) (Merck, catalog number: 1.04936.1000)
Sodium phosphate dibasic (Na2HPO4) (Merck, catalog number: S9763)
Potassium phosphate monobasic (KH2PO4) (Merck, catalog number: P0662)
Hydrochloric acid (HCl) (Merck, catalog number: 1.00317.2500)
NH4Cl (Merck, catalog number: 254134)
Colorless nail polish
GalT-mCherry mammalian expression plasmid DNA (Tie et al., 2016)
GFP-OSBP mammalian expression plasmid DNA (Addgene: pLJM1-FLAG-GFP-OSBP #134659)
Primary antibodies
GM130 mouse monoclonal antibody (BD Biosciences, catalog number: 610823; dilution 1:500)
Giantin rabbit polyclonal antibody (BioLegend, catalog number: 924302; dilution 1:1,000)
Secondary antibodies
Goat anti-mouse IgG conjugated with Alexa Fluor (AF) 647 (Invitrogen, catalog number: A21235; dilution 1:500)
Goat anti-rabbit IgG conjugated with AF488 (Invitrogen, catalog number: A11008; dilution 1:500)
Goat anti-rabbit IgG conjugated with AF647 (Invitrogen, catalog number: A32733; dilution 1:500)
Solutions
33 mM nocodazole stock solution (see Recipes)
Phosphate buffered saline (PBS) (see Recipes)
1 M Tris pH 8.0 (see Recipes)
4% paraformaldehyde fixative (see Recipes)
100 mM NH4Cl (see Recipes)
10% saponin (see Recipes)
Antibody dilution buffer (see Recipes)
Mowiol mounting medium (see Recipes)
Equipment
A pair of sharp forceps
Water bath
Airyscan microscope: Zeiss LSM980 system equipped with a motorized stage, a temperature-controlled environment chamber, 63×/1.46 Oil Plan-Apochromat objective, and Zeiss Airyscan 2 detector module. GFP or AF488, mCherry, and AF647 are excited by 488, 561, and 639 nm diode laser lights, respectively. The emission lights from GFP or AF488, mCherry, and AF647 are collected by BP495-560, BP570-630, and LP655 filters.
37 °C cell culture incubator with 5% CO2 (Nuaire, catalog number: NU5510E)
Benchtop centrifuge (Eppendorf, catalog number: EPPE5401000.064)
Software
Fiji (NIH, https://fiji.sc/)
Macros (Supplementary Files 1–6): P1-Rotate_Resize_Normalize, P2-Resize_Add_Line, P3-Reflection_Average, ROI_to_image, Side average_Axial line intensity profile, and Side average_Lateral line intensity profile.
Zeiss Zen software
Microsoft Excel
Microsoft Word
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Mahajan, D., Tie, H. C. and Lu, L. (2023). Visualizing the Cisternal Organization of Golgi Ministacks in HeLa Cells by Side-averaging. Bio-protocol 13(8): e4658. DOI: 10.21769/BioProtoc.4658.
- Tie, H. C., Mahajan, D. and Lu, L. (2022). Visualizing intra-Golgi localization and transport by side-averaging Golgi ministacks. J Cell Biol 221(6): e202109114.
分类
细胞生物学 > 细胞成像 > 荧光
细胞生物学 > 细胞成像 > 超分辨率成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link