- EN - English
- CN - 中文
Unscheduled DNA Synthesis at Sites of Local UV-induced DNA Damage to Quantify Global Genome Nucleotide Excision Repair Activity in Human Cells
在局部紫外线诱导的 DNA 损伤位点进行程序外DNA合成,以量化人类细胞中的全基因组核苷酸切除修复活性
发布: 2023年02月05日第13卷第3期 DOI: 10.21769/BioProtoc.4609 浏览次数: 2222
评审: Alak MannaAnonymous reviewer(s)
Abstract
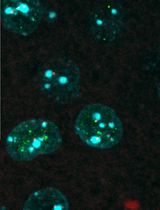
Nucleotide excision repair (NER) removes a wide variety of structurally unrelated lesions from the genome, including UV-induced photolesions such as 6–4 pyrimidine–pyrimidone photoproducts (6-4PPs) and cyclobutane pyrimidine dimers (CPDs). NER removes lesions by excising a short stretch of single-stranded DNA containing the damaged DNA, leaving a single-stranded gap that is resynthesized in a process called unscheduled DNA synthesis (UDS). Measuring UDS after UV irradiation in non-dividing cells provides a measure of the overall NER activity, of which approximately 90% is carried out by the global genome repair (GGR) sub pathway. Here, we present a protocol for the microscopy-based analysis and quantification of UDS as a measurement for GGR activity. Following local UV-C irradiation, serum-starved human cells are supplemented with the thymidine analogue 5-ethynyl-2'-deoxyuridine (EdU), which is incorporated into repair patches following NER-dependent dual incision. The incorporated nucleotide analogue is coupled to a fluorophore using Click-iT chemistry, followed by immunodetection of CPD photolesions to simultaneously visualize both signals by fluorescence microscopy. Accompanying this protocol is a custom-built ImageJ plug-in to analyze and quantify UDS signals at sites of CPD-marked local damage. The local UDS assay enables an effective and sensitive fluorescence-based quantification of GGR activity in single cells with application in basic research to better understand the regulatory mechanism in NER, as well as in diagnostics to characterize fibroblasts from individuals with NER-deficiency disorder.
Graphical abstract

Background
Cells are continuously exposed to different sources of DNA damage. The nucleotide excision repair (NER) pathway comprises two sub pathways, transcription-coupled repair (TCR) and global genome repair (GGR), which remove helix-distorting lesions including UV-induced photoproducts from genomic DNA. While TCR preferentially removes DNA lesions from the transcribed strand of active genes (van den Heuvel et al., 2021), GGR removes lesions throughout the genome and is the dominant contributor to overall cellular NER activity (Marteijn et al., 2014). Both pathways funnel into a common molecular mechanism involving the same set of core NER proteins that mediate double incision by endonucleases ERCC1-XPF and XPG, resulting in the removal of a 30-nucleotide stretch of single-stranded DNA containing the lesion (van Toorn et al., 2022). After dual incision, the resulting gap is filled by DNA polymerases and sealed by DNA ligases (Ogi et al., 2010). DNA repair–associated DNA synthesis is distinct from synthesis during the S-phase to duplicate the genome, and is therefore referred to as unscheduled DNA synthesis (UDS) (Lehmann and Stevens, 1980). Irradiation with UV-C light generates two main types of genomic lesions: 6–4 pyrimidine–pyrimidone photoproducts (6-4PPs) and cyclobutane pyrimidine dimers (CPDs). The 6-4PPs are repaired considerably faster (within ∼5 h) than the CPDs, which are still present 24 h after UV irradiation (van Hoffen et al., 1995; Luijsterburg et al., 2010; Verbruggen et al., 2014). UDS after UV irradiation can be directly measured via nucleotide incorporation in non-dividing cells and mainly reflects GGR activity, which is responsible for approximately 90% of the overall cellular NER activity (Limsirichaikul et al., 2009). Although radioactive nucleotide analogues were initially used to measure UDS (Cleaver, 1968; Friedberg, 2004), the use of alkyne-linked thymidine analogue 5-ethynyl-2'-deoxyuridine (EdU) is more common nowadays (Limsirichaikul et al., 2009). The incorporated EdU can be visualized by its coupling to a fluorescent azide via Click-iT chemistry (Salic and Mitchison, 2008). Measuring UDS can offer insight in the potential involvement of GGR regulators or assess the functionality of mutated NER proteins, as recently reported for chromatin remodeler ALC1 or the ERCC1R156W variant found in patients with liver and kidney dysfunction, respectively (Blessing et al., 2022;Apelt et al., 2021). In this protocol, we describe a straightforward and accessible method for measuring UDS at sites of local UV irradiation. In brief, cells are exposed to local UV-C irradiation through a micropore filter, causing DNA damage induction and NER-dependent UDS in sub-nuclear regions. The UDS signal at sites of local UV damage can be captured by pulse-labeling with EdU within the first few hours after UV irradiation, followed by fixation and visualization of the incorporated EdU by fluorescent Click-iT chemistry. NER-dependent DNA synthesis mainly reflects rapid 6-4PP repair within the first hours after UV irradiation, during which CPD repair is negligible (van Hoffen et al., 1995; Luijsterburg et al., 2010; Verbruggen et al., 2014). The simultaneous immunodetection of CPD photolesions, which are largely unrepaired within the first hours after UV irradiation, can be used to define sites of local damage. The use of local UV irradiation enables a per-cell correction for unspecific EdU incorporation and accurate exclusion of replicating cells with scheduled EdU incorporation throughout the nucleus. Moreover, we provide a custom-built ImageJ plug-in to quantify UDS signals and apply a correction for potential differences in DNA damage induction between experimental conditions based on the CPD signal. We show that local UDS at sites of UV-induced DNA damage is robustly detected in RPE1-hTERT human cells, while the UDS signal is nearly abolished in an isogenic knockout cell-line for essential NER geneXPG.
Materials and Reagents
12-well cell culture plate (Sigma, catalog number: CLS3512)
18 mm cover glasses, round (VWR, catalog number: 631-1580)
5 µm polycarbonate filters (Millipore, catalog number: TMTP04700)
Parafilm (Sigma, catalog number: P7543)
Microscope slides (VWR, catalog number: 12362098)
DMEM, high glucose, GlutaMAXTM supplement, pyruvate (Gibco, catalog number: 31966-047)
Fetal bovine serum (FBS) (Capricorn Scientific, catalog number: FBS-12A)
5-Ethynyl-2'-deoxyuridine (5-EdU) (Jena Bioscience, catalog number: CLK-N001-100); 50 mM stock in PBS [100 mg in 7.9 mL; to dissolve put in water bath (70°C) for 1 min (until dissolved); aliquot and store at -20°C]
5-Fluoro-2’-deoxyuridine (FuDR) (Sigma, catalog number: F0503); 10 mM stock in MilliQ water (MQ) [100 mg in 40.6 mL; aliquot and store at -20°C]
Thymidine (Sigma, catalog number: T1895); 100 mM stock in MQ [1 g in 41.3 mL; aliquot and store at -20°C]
Formaldehyde 37% (Sigma, catalog number: 252549)
Triton-X100 (Sigma, catalog number: X100)
Bovine serum albumin fraction V (BSA) (Sigma, catalog number: 10735086001)
Tris (Sigma, catalog number: 10708976001)
Atto azide Alexa 647 (Atto Tec, catalog number: 647N-101); 6 mM stock in DMSO [1 mg in 173 µL; aliquot and store at -20°C]
CuSO4 (VWR, catalog number: 23174.233); 40 mM stock in MQ [store at room temperature (RT)]
Ascorbic acid (Sigma, catalog number: A0278); 20 mg/mL stock in MQ [prepare fresh before use]
Glycine (Sigma, catalog number: G7126)
NaOH (Sigma, catalog number: 567530)
Tween20 (Sigma, catalog number: P1379)
CPD antibody (Cosmo Bio, CAC-NM-DND-001)
Goat anti-mouse Alexa 555 (Thermo Fisher Scientific, catalog number: A-21424)
DAPI (Thermo Fisher Scientific, catalog number: D1306)
Aqua poly/mount (Brunschwig, catalog number: 18606)
Sterile PBS (Thermo Fisher Scientific, catalog number: 10010023)
Click reaction mix (see Recipes)
Wash buffer (WB) (see Recipes)
Equipment
Tweezers (stainless steel forceps curved, Sigma, Z168785)
TUV PL-S 9 W UV-C lamp (Philips)
International Light NIST traceable radiometer photometer (Model IL1400BL SEL240/NS254/W)
Fluorescence microscope (Zeiss Axio Imager M2)
Software
Imaging software: Zen (2012 Blue edition)
Data analyses software: ImageJ (version 1.48), with the following requirements:
ImageJ Plugin “bioformats_package.jar” at https://www.openmicroscopy.org/bio-formats/downloads
In-house generated ImageJ macro “GGR-UDS_DataExtraction_ImageJ.txt” available at https://git.lumc.nl/dvandenheuvel/ggr-uds-protocol_lumc_luijsterburglab
In-house generated data quantification file GGR-UDS_analyses_template.xls available at https://git.lumc.nl/dvandenheuvel/ggr-uds-protocol_lumc_luijsterburglab
Procedure
文章信息
版权信息
© 2023 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
van der Meer, P., van den Heuvel, D. and Luijsterburg, M. S. (2023). Unscheduled DNA Synthesis at Sites of Local UV-induced DNA Damage to Quantify Global Genome Nucleotide Excision Repair Activity in Human Cells. Bio-protocol 13(3): e4609. DOI: 10.21769/BioProtoc.4609.
分类
分子生物学 > DNA > DNA 损伤和修复
细胞生物学 > 细胞成像 > 荧光
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link