- EN - English
- CN - 中文
Assay for Phytaspase-mediated Peptide Precursor Cleavage Using Synthetic Oligopeptide Substrates
使用合成寡肽底物测定植物蛋白酶介导的肽前体裂解
发布: 2023年02月05日第13卷第3期 DOI: 10.21769/BioProtoc.4608 浏览次数: 1794
评审: Giusy TornilloBilly Tasker-BrownSimab Kanwal
Abstract
Proteases control plant growth and development by limited proteolysis of regulatory proteins at highly specific sites. This includes the processing of peptide hormone precursors to release the bioactive peptides as signaling molecules. The proteases involved in this process have long remained elusive. Confirmation of a candidate protease as a peptide precursor–processing enzyme requires the demonstration of protease-mediated precursor cleavage in vitro. In vitro cleavage assays rely on the availability of suitable substrates and the candidate protease with high purity. Here, we provide a protocol for the expression, purification, and characterization of tomato (Solanum lycopersicum) phytaspases as candidate proteases for the processing of the phytosulfokine precursor. We also show how synthetic oligopeptide substrates can be used to demonstrate site-specific precursor cleavage.
Graphical abstract
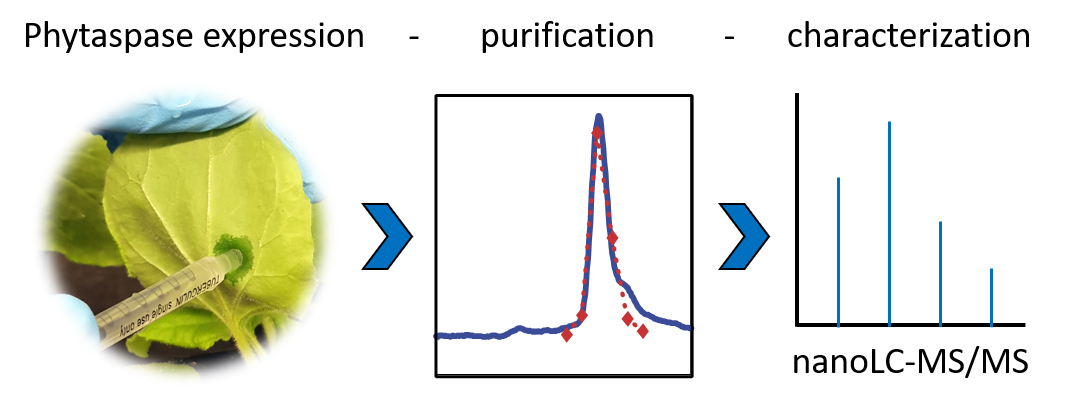
Background
Phtytaspases are a subgroup of plant subtilases that are characterized by their specificity for aspartic acid immediately upstream of the cleavage site (in position P1) of their oligopeptide and protein substrates (Chichkova et al., 2014; Schaller et al., 2018). Phytaspases were originally characterized in tobacco and rice (Chichkova et al., 2010) and more recently in Arabidopsis and tomato (Beloshistov et al., 2018; Chichkova et al., 2018; Reichardt et al., 2018 and 2020). In addition to the canonical Asp residue at the scissile bond, several amino acids upstream of the critical Asp were found to contribute to substrate recognition, resulting in high selectivity of individual phytaspases for limited proteolysis at specific sites of their protein targets (Chichkova et al., 2018; Reichardt et al., 2018).
Phytaspases in particular, and plant subtilases in general, are synthesized as pre-pro-proteins that are directed to the secretory pathway for proteolytic maturation, glycosylation, and secretion (Schaller et al., 2018). While there is a single report on the successful expression of phytaspase in E. coli (Narayanan et al., 2017), the complex maturation pattern of subtilases rather calls for an eukaryotic host for recombinant protein expression (Meyer et al., 2016). The baculovirus–insect cell system and stably transformed plant cell lines have been used to produce post-translationally modified and fully active plant subtilases (Janzik et al., 2000; Cedzich et al., 2009; Ottmann et al., 2009). Here, we used Nicotiana benthamiana plants for the expression of C-terminally His-tagged phytaspases by agroinfiltration. This transient expression system may not yield as much recombinant protein as insect and plant cell culture systems but is much more rapid and allows for simple extraction of the secreted subtilases from extracellular (apoplastic) wash fluids. Subsequent purification by metal-chelate affinity chromatography followed by gel filtration results in near homogeneity of the recombinant enzymes for in vitro cleavage assays.
Cleavage assays include synthetic oligopeptides that comprise several precursor-derived amino acids upstream and/or downstream of the cleavage sites as protease substrates. In this experiment, we used a decamer peptide consisting of five amino acids of the precursor followed by the di-sulfated PSK pentapeptide as a substrate for tomato phytaspases. To demonstrate specificity of cleavage, we included a second peptide, in which Ala substituted the critical Asp residue at the cleavage site. Cleavage products were then identified and quantified by mass spectrometry (Reichardt et al., 2020).
We provide a protocol for (A) the transient expression of His-tagged phytaspases by agroinfiltration of N. benthamiana plants, (B) the purification of the recombinant proteins from apoplastic leaf extracts, (C) the cleavage assay using synthetic peptide substrates, and (D) sample preparation for mass spectrometry (MS). Not included is a protocol for the cloning of the protease of interest. In the experiment described here, we used expression constructs for phytaspases from tomato that were generated by conventional cloning techniques in the binary vector pART27 and transformed into Agrobacterium tumefaciens strain C58C1 as described (Reichardt et al., 2018). A protocol for the mass spectrometric analysis of cleavage products is also not included, as this part of the analysis is usually either performed by a central facility of the respective institution or provided as a commercial service. For SDS-PAGE analysis, we followed standard procedures (Stintzi et al., 2022). The protocols we provide here can easily be adapted to other secreted proteases that tolerate the addition of a C-terminal His tag. Peptide sequences will have to be chosen according to the specific substrate requirements of the protease of interest.
Materials and Reagents
50 mL culture tubes (Corning/Falcon, catalog number: 352070)
15 mL culture tubes (Corning/Falcon, catalog number: 352196)
1 mL PE/PP syringes (avantor/VWR, catalog number: 613-2001)
100 mL PE/PP syringes (Th.Geyer/Becton Dickinson, catalog number: 6287774)
Oak Ridge High-Speed PPCO centrifuge tubes (Thermo Fisher Scientific/Nalgene, catalog number 3139-0030)
250 or 500 mL centrifuge bottles (Thermo Fisher Scientific/Nalgene, catalog number: 3141-0250 or 3141-0500)
Scalpel or razor blades
17 or 18 gauge blunt-tipped syringe needle (e.g., B. Braun Sterican®, 18G / 1.2 × 40 mm, catalog number: 4038088)
300 or 500 mL Pyrex beakers
Vivaspin 20 centrifugal concentrator (30 k molecular weight cut-off) (Sartorius Stedim, catalog number: VS2021)
A. tumefaciens C58C1 (RifR, TetR) (community resource; NCBI:txid176299)
Expression construct for the protease of interest under control of the CaMV 35S promoter in a binary vector for plant transformation. Here, we used expression constructs for His-tagged tomato phytaspases in pART27 (SpecR) transformed into A. tumefaciens, strain C58C1 (Reichardt et al., 2018). Agrobacteria transformed with the empty expression vector (here pART27) were used as control
A. tumefaciens C58C1 with p19 silencing suppressor in pBin61 (KanR, RifR, TetR) (Voinnet et al., 2003)
Nicotiana benthamiana seeds (Agroscience GmbH, Neustadt, Germany)
N. benthamiana plants, grown on seeding substrate at 25°C and 16:8 h day/night cycle. Plants older than three weeks are watered with 1.48 g/L N (20%), P (20%), and K (20%) universal fertilizer including micronutrients.
Note: Expression levels are much reduced in flowering plants. Therefore, use the plants before they start to bolt, usually when they are between four and five weeks old
Spectinomycin (Duchefa, catalog number: S0188); 100 mg/mL stock solution in H2O, store at -20°C
Rifampicin (Duchefa, catalog number: R0146); 100 mg/mL stock solution in DMSO, store at -20°C
Tetracycline (Duchefa, catalog number: T0150); 25 mg/mL stock solution in 70 % (v/v) ethanol, store at -20°C
Kanamycin sulphate (Duchefa, catalog number: K0126); 50 mg/mL stock solution in H2O, store at -20°C
Tryptone (Duchefa, catalog number: T1332)
Nickel (Ni)-NTA agarose (Qiagen, catalog number: 30210), store at 4°C
Bio-Rad Protein Assay kit II, with bovine serum albumin as the standard protein (Bio-Rad, catalog number: 5000002)
Coomassie protein stain (e.g., InstantBlue, abcam, catalog number: ISB1L)
Custom-synthesized synthetic substrate peptides at >90% purity, EAHLD[sY]I[sY]TQM and EAHLA[sY]I[sY]TQM (sY = sulfotyrosine), (PepMic, Suzhou, China). The lyophilized peptides are stored at -20°C. Working solutions can be stored at 4°C for one month, or at -20°C. Avoid repeated freeze/thaw cycles
MgCl2·6H2O (Carl Roth, catalog number: 3532.1)
NaCl (Carl Roth, catalog number: 9265.2)
KCl (Carl Roth, catalog number: 6781.1)
NaH2PO4·2H2O (Carl Roth, catalog number: T879.2)
Na2HPO4·2H2O (Carl Roth, catalog number: T877.1)
NaOH (Carl Roth, catalog number: P031.3)
Acetosyringone (Carl Roth, catalog number: 6003.2), store at -20°C
Imidazole (Carl Roth, catalog number: 3899.4)
2-(N-Morpholino)-ethanesulfonic acid (MES·H2O) (Carl Roth, catalog number: 6066.2)
Yeast extract (Carl Roth, catalog number: 2904.3)
Glacial acetic acid (Carl Roth, catalog number: 7332.1)
Glycerol (Carl Roth, catalog number: 4043.1)
Acetonitrile (Carl Roth, catalog number: 7330.2)
Trifluoroacetic acid (Carl Roth, catalog number: P088.1)
H2O, HPLC grade (Carl Roth, catalog number: A511.2)
LB medium (lysogeny broth) (see Recipes)
Infiltration buffer (see Recipes)
Extraction buffer, reaction buffer (see Recipes)
Binding buffer (see Recipes)
Elution buffer (see Recipes)
Gel filtration buffer (see Recipes)
Solvent A (see Recipes)
Solvent B (see Recipes)
Solvent C (see Recipes)
Equipment
Microcentrifuge (Eppendorf, model: 5418 R)
Tabletop centrifuge with swing-out rotor for 15/50 mL tubes (Eppendorf, model: 5810 R)
RC-3B refrigerated centrifuge (Sorvall Instruments) for 250/500 mL bottles
RC6+ refrigerated centrifuge (Sorvall Instruments) for 30 mL tubes
UV/Vis spectrophotometer (e.g., Eppendorf Biophotometer, catalog number: 634-0839)
Desiccator (5 L) with vacuum pump
Rotating wheel (e.g., Steinberg Systems, model: SBS-LBM-200)
Microbiological incubators for plate cultures at 28 and 37°C
Enrich SEC 650 10 × 300 gel filtration column (Bio-Rad, catalog number: 7801650)
Fast protein liquid chromatography system. We used the NGC Quest system (Bio-Rad, catalog number: 788-0003)
Note: Other chromatography systems, e.g., ÄKTA Pure (Cytiva, catalog number: 29018226) can be used as well.
Vacuum concentrator, e.g., Savant SpeedVac (Thermo Fisher Scientific, catalog number: SPD1030A)
Standard SDS-PAGE equipment (e.g., Mini-PROTEAN Tetra Cell and casting module from Bio-Rad, catalog numbers: 1658000EDU and 1658015EDU)
Electrophoresis power supply (e.g., PowerPac Basic from Bio-Rad, catalog number: 1645050)
PTFE (polytetrafluorethylene; TeflonTM) membranes with embedded C18 beads, e.g., Supelco EmporeTM solid phase extraction discs (Millipore Sigma/Supelco, catalog number: 66887U)
Procedure
文章信息
版权信息
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Reichardt, S., Stintzi, A. and Schaller, A. (2023). Assay for Phytaspase-mediated Peptide Precursor Cleavage Using Synthetic Oligopeptide Substrates. Bio-protocol 13(3): e4608. DOI: 10.21769/BioProtoc.4608.
- Reichardt, S., Piepho, H.-P., Stintzi, A. and Schaller, A. (2020). Peptide signaling for drought-induced tomato flower drop. Science 367(6485): 1482-1485.
分类
植物科学 > 植物生物化学 > 蛋白质
生物化学 > 蛋白质 > 合成
生物化学 > 蛋白质 > 分离和纯化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link