- EN - English
- CN - 中文
Isolation of Nuclei from Human Snap-frozen Liver Tissue for Single-nucleus RNA Sequencing
从人速冻肝组织中分离细胞核用于单核 RNA 测序
发布: 2023年02月05日第13卷第3期 DOI: 10.21769/BioProtoc.4601 浏览次数: 2588
评审: Laxmi Narayan MishraNitin AmdareAmit Kumar
Abstract
Single-nucleus RNA sequencing (snRNA-seq) provides a powerful tool for studying cell type composition in heterogenous tissues. The liver is a vital organ composed of a diverse set of cell types; thus, single-cell technologies could greatly facilitate the deconvolution of liver tissue composition and various downstream omics analyses at the cell-type level. Applying single-cell technologies to fresh liver biopsies can, however, be very challenging, and snRNA-seq of snap-frozen liver biopsies requires some optimization given the high nucleic acid content of the solid liver tissue. Therefore, an optimized protocol for snRNA-seq specifically targeted for the use of frozen liver samples is needed to improve our understanding of human liver gene expression at the cell-type resolution. We present a protocol for performing nuclei isolation from snap-frozen liver tissues, as well as guidance on the application of snRNA-seq. We also provide guidance on optimizing the protocol to different tissue and sample types.
Keywords: Nuclei isolation (细胞核分离)Background
The human liver performs critical functions ranging from lipid metabolism, amino acid synthesis, and drug processing to protection from portal venous bacteria and viruses (Trefts et al., 2017). Consistent with this diverse range of functions, its spatial organization is specifically tailored (Ben-Moshe and Itzkovitz, 2019). For example, lobule zonation between a central vein and portal triad divides hepatocyte and endothelial functions (Halpern et al., 2017). Diverse and specialized cell types populate the liver (Aizarani et al., 2019), and thus studies involving the liver would benefit greatly from single-cell resolution.
The field of single-cell genomics has accelerated cellular composition studies of tissues and samples. Single-cell RNA sequencing (RNA-seq) has been used to discover subtypes within blood monocytes and dendritic cells (Villani et al., 2017), as well as to discover myeloid differentiation pathways (Drissen et al., 2016). In solid tissues, single-cell RNA-seq has provided insight into fibrosis mechanisms in lung (Xu et al., 2016) and kidney (Kuppe et al., 2021). Single-cell RNA-seq has also been used in liver to transcriptionally characterize the landscape of cell types (MacParland et al., 2018), as well as the immune landscape in hepatocellular carcinoma (HCC) (Zhang et al., 2019).
One adaptation of single-cell RNA-seq is the use of nuclei instead of whole cells. Single-nucleus RNA-seq (snRNA-seq) has been shown to produce similar results as scRNA-seq (Habib et al., 2017). Furthermore, it can identify a greater expression diversity within cell types than scRNA-seq (Andrews et al., 2022). A main advantage of snRNA-seq is its applicability to frozen tissues. Archived tissues remove the need to coordinate tissue processing immediately after biopsy collection, which is challenging for human tissues. Archived tissues may also have additional phenotype and molecular data readily available, allowing for more thorough analyses and controlling of confounders.
However, the isolation of nuclei from frozen tissues for snRNA-seq presents distinct challenges. Snap freezing can result in the formation of ice crystals, disrupting the tissue and possibly decreasing the yield of intact nuclei (Larson and Chin, 2021). This can also result in higher amounts of tissue debris and contaminating ambient RNA in the lysate and nucleus suspension. We present a specifically tailored protocol for snRNA-seq of human frozen liver tissue (Rao et al., 2021), balancing requirements for yield and debris removal. For an overview of the protocol, see Figure 1. The advantage of our protocol is the rapid isolation of nuclei with minimum equipment, which prevents RNA degradation and increases throughput. Our protocol avoids labor-intensive flow cytometry sorting, which may require high yields and decrease throughput. We also avoid density-based centrifugation, which increases run times and susceptibility to RNA degradation. In addition, we provide details on how to isolate nuclei suitable for droplet-based microfluidics applications, as well as data analysis guidelines. While optimized for snap-frozen liver tissue, this protocol can be adapted to other frozen tissue samples, such as adipose tissue (Alvarez et al., 2020).
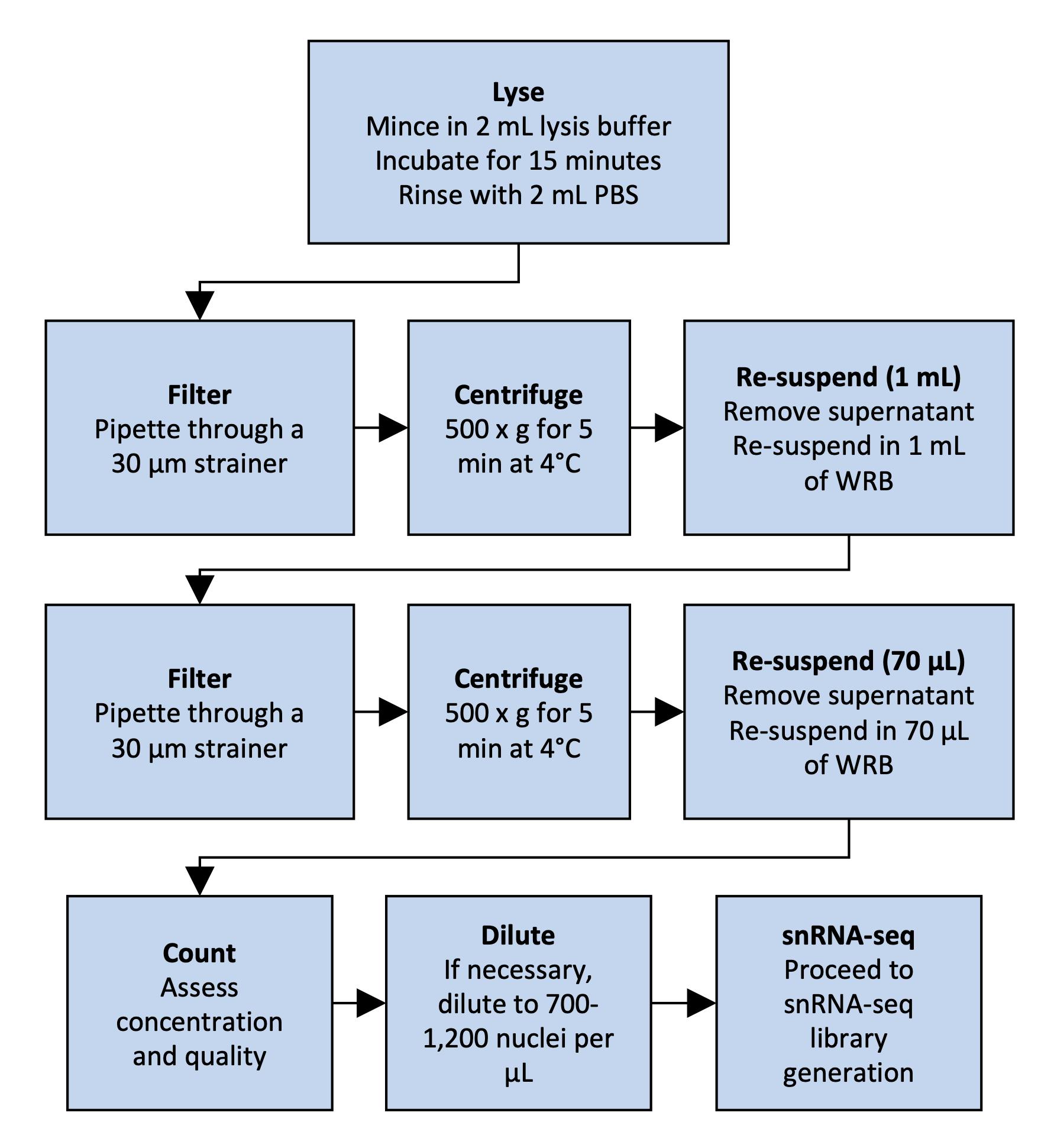
Figure 1. Overview of nucleus isolation from frozen liver. The flowchart outlines the steps from tissue lysis to nuclear suspension.
Materials and Reagents
100 × 15 mm Petri dish (Corning®, Falcon®, catalog number: 351029)
Single-use scalpels No. 10 (Fisher Scientific, FeatherTM, catalog number: 08-927-5A)
MACS® SmartStrainers 30 μm (Miltenyi Biotec, catalog number: 130-110-915)
FlowMiTM 40 μm pipette tip strainers (Bel-Art, catalog number: H13680-0040)
15 mL FalconTM conical centrifuge tubes (Corning®, catalog number: 352096)
50 mL FalconTM conical centrifuge tubes (Corning®, catalog number: 352070)
DNA LoBind® 1.5 mL microcentrifuge tubes (Eppendorf, catalog number: 022431021)
FisherbrandTM tweezers/forceps (Fisher Scientific, catalog number: 12-000-128)
Countess cell counting chamber slides (Thermo Fisher Scientific, catalog number: C10228)
Phosphate-buffered saline (PBS), 1× without calcium and magnesium, pH 7.4 (Corning®, catalog number: 21-040-CM)
Nuclease-free water (not DEPC-treated) (Fisher Scientific, InvitrogenTM, catalog number: AM9932)
IGEPAL® CA-630 (Millipore Sigma, catalog number: 18896-50ML)
Sodium chloride (NaCl) (Millipore Sigma, catalog number: S5886-500G)
Magnesium chloride (MgCl2) (Millipore Sigma, catalog number: M2393-100G)
Bovine serum albumin (BSA) (Millipore Sigma, catalog number: A8806-5G)
Hoechst 33342 10 mg/mL solution (Thermo Fisher Scientific, catalog number: H3570)
Protector RNase inhibitor (Millipore Sigma, catalog number: 3335402001)
Trizma® hydrochloride solution (Tris-HCl) 1 M pH 7.4 (Millipore Sigma, catalog number: T2194)
5 M NaCl stock (see Recipes)
1 M MgCl2 stock (see Recipes)
10% IGEPAL (see Recipes)
0.1% lysis buffer (see Recipes)
Wash and resuspension buffer (WRB) (see Recipes)
Hoechst stain buffer (see Recipes)
Equipment
Refrigerated benchtop centrifuge
Note: Using a swinging bucket rotor for centrifugation helps to improve nuclei yields when compared with a fixed-angle rotor centrifuge.
Countess II FL automated cell counter, or equivalent cell counting method
Agilent Bioanalyzer
Procedure
文章信息
版权信息
© 2023 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Alvarez, M., Benhammou, J. N., Rao, S., Mishra, L., Pisegna, J. R. and Pajukanta, P. (2023). Isolation of Nuclei from Human Snap-frozen Liver Tissue for Single-nucleus RNA Sequencing. Bio-protocol 13(3): e4601. DOI: 10.21769/BioProtoc.4601.
- Rao, S., Yang, X., Ohshiro, K., Zaidi, S., Wang, Z., Shetty, K., Xiang, X., Hassan, M. I., Mohammad, T., Latham, P. S., et al. (2021). beta2-spectrin (SPTBN1) as a therapeutic target for diet-induced liver disease and preventing cancer development. Sci Transl Med 13(624): eabk2267.
分类
分子生物学 > RNA > RNA 测序
细胞生物学 > 细胞器分离 > 细胞核
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link