- EN - English
- CN - 中文
Arrayed CRISPR/Cas9 Screening for the Functional Validation of Cancer Genetic Dependencies
用于癌症遗传依赖性功能验证的阵列式 CRISPR/Cas9 筛选
发布: 2022年12月20日第12卷第24期 DOI: 10.21769/BioProtoc.4577 浏览次数: 3078
评审: Rene WinklerSalim GasmiAnonymous reviewer(s)

相关实验方案
在哺乳动物细胞中通过具有 3' 突出端的长 dsDNA 介导的 CRISPR 敲入 (LOCK) 实现高效的大 DNA 片段敲入
Wenjie Han [...] Jianqiang Bao
2023年10月20日 2770 阅读
Abstract
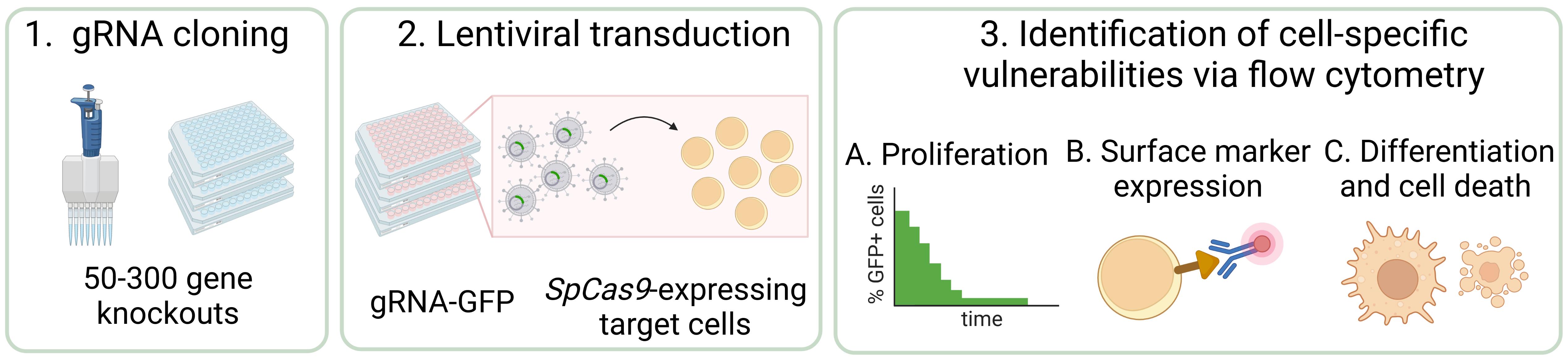
CRISPR/Cas9 screening has revolutionized functional genomics in biomedical research and is a widely used approach for the identification of genetic dependencies in cancer cells. Here, we present an efficient and versatile protocol for the cloning of guide RNAs (gRNA) into lentiviral vectors, the production of lentiviral supernatants, and the transduction of target cells in a 96-well format. To assess the effect of gene knockouts on cellular fitness, we describe a competition-based cell proliferation assay using flow cytometry, enabling the screening of many genes at the same time in a fast and reproducible manner. This readout can be extended to any parameter that is accessible to flow-based measurements, such as protein expression and stability, differentiation, cell death, and others. In summary, this protocol allows to functionally assess the effect of a set of 50–300 gene knockouts on various cellular parameters within eight weeks.
Graphical abstract
Background
The experimental identification of genes that are essential for cancer cell proliferation and survival has radically changed over the last decade. The CRISPR/Cas9 technology is a powerful tool for the assessment of cancer-specific vulnerabilities (u and Yusa, 2019; Behan et al., 2019). It has been successfully employed in various cancer models and led to the identification of multiple genetic dependencies, such as WRN1 in cancers with microsatellite instability, YTHDF2 in triple-negative breast cancer, TRIM8 in Ewing sarcoma, or STAUFEN2 and ERG in acute myeloid leukemia (Lin and Sheltzer, 2020; Chan et al., 2019; Bajaj et al., 2020; Einstein et al., 2021; Seong et al., 2021; Schmoellerl et al., 2022). Indeed, pooled genome-wide loss-of-function screens to identify genetic dependencies have become common practice in cancer biology (Behan et al., 2019; Bajaj et al., 2020) as well as in many other fields of the molecular life sciences, such as stem cell biology, immunology, or cell and developmental biology (Arroyo et al., 2016; Y. Zhu, et al., 2021; Dong et al., 2022; Zhang et al., 2022). In parallel, pooled focused screens covering smaller sets of genes representing specific pathways or protein families are often employed (X. G. Zhu et al., 2021; Cao et al., 2021). Furthermore, next-generation sequencing–based methods, such as RNA-seq, ATAC-seq, or Cut&Run, as well as mass spectrometry–based proteomics are increasingly used to characterize molecular alterations in cancer cells in an unbiased global fashion. However, all these experimental approaches yield large amounts of data and high numbers of candidate genes, ranging from a few hundred to several thousand. Consequently, streamlined strategies for efficient functional genomic screening and validation of candidate genes downstream of pooled screens need to be developed. Here, we describe how to design and perform arrayed CRISPR/Cas9 screening experiments of candidate sets ranging from 50 to 300 gene knockouts for the validation and identification of functional genetic dependencies of cancer cells using flow cytometry that can serve as starting points for detailed molecular analyses.
Materials and Reagents
Cloning
Scalpel
Pipette tips, filtered, sterile, 0.5–1,250 µL (Biozym Surphob)
1.5 mL tubes (Eppendorf, catalog number: 0030121872)
96-well PCR plates (Biozym Scientific, catalog number: 710880)
96-well deep well plates, 2.2 mL, V-bottom (Biozym Scientific, catalog number: 710850)
LentiGuide-Puro-P2A-EGFP (Addgene, Plasmid #137729) (Panda et al., 2020)
BsmBI-v2 (New England Biolabs, catalog number: R0739L)
NEBuffer r3.1 (New England Biolabs, catalog number: B6003S)
Double-distilled water (ddH2O)
Antarctic phosphatase (New England Biolabs, catalog number: M0289L)
Antarctic phosphatase reaction buffer (New England Biolabs, catalog number: B0289S)
Agarose (Sigma Aldrich, catalog number: A9539)
Monarch® DNA Gel Extraction kit (New England Biolabs, catalog number: T1020L)
T4 DNA ligase (New England Biolabs, catalog number: M0202L)
T4 DNA ligase reaction buffer (New England Biolabs, catalog number: B0202S)
T4 polynucleotide kinase (PNK) (New England Biolabs, catalog number: M0201L)
NEB stable competent E. coli (New England Biolabs, catalog number: C3040H)
LB medium (Carl Roth, catalog number: X964.4)
LB agar (Carl Roth, catalog number: 6671.2)
Carbenicillin (Carl Roth, catalog number: 6344)
Petri dish, square, 12 × 12 cm (Greiner Bio-One, catalog number: 688161)
Inoculation loop (Sarstedt, catalog number: 86.1562.050)
Monarch® Plasmid Miniprep kit (New England Biolabs, catalog number: T1010L)
Cell culture
Sterile flat-bottom 96-well plates (Greiner, catalog number: 655180)
Round bottom 96-well plates (Greiner, catalog number: 650188)
10 cm2 dishes (Greiner, catalog number: 655160)
24-well cell culture plates (Stölzle Oberglas, catalog number: GR-662160)
Syringe filter 0.45 µm, 33 mm diameter (M&B Stricker, catalog number: 99745)
Sterile distilled water (Ecotainer, Braun, catalog number: 82479E-E)
DMEM high glucose, no glutamine, 10 × 500 mL (Life Tech, catalog number: 11960085)
Fetal bovine serum (FBS), heat inactivated (Sigma-Aldrich, catalog number: F9665-500ML)
L-glutamine 200 mM (100×), liquid (Gibco, catalog number: 25030024)
Penicillin-streptomycin, 10,000 U/mL (Life Tech, catalog number: 15140122)
Phosphate buffered saline (PBS), 10×, pH 7.2–10 (Life Tech, catalog number: 70013065)
Trypsin, 0.25% (1×) with EDTA 4Na, liquid (Gibco, catalog number: 25200056)
Lenti-X 293T cells (Takara bio, catalog number: 632180)
psPAX2 (packaging plasmid) (Addgene, catalog number: 12260)
pMD2.G (packaging plasmid) (Addgene, catalog number: 12259)
Polyethylenimine HCl MAX, linear, MW 40,000 (PEI) (Polysciences Europe, catalog number: 24765-1)
PEI stock: 1 mg/mL in water according to the supplier’s protocol
Polybrene transfection reagent (Merck, catalog number: TR-1003-G)
Oligonucleotides
DNA oligonucleotide primers from Integrated DNA Technologies (IDT) in 96-well format, premixed
hU6 prom_fwd GACTATCATATGCTTACCGT
Polybrene stock: 10 mg/mL in sterile water
Equipment
Single-channel pipettes, 0.5–1000 µL (Eppendorf Research)
Multi-channel pipettes, 1–200 µL (Thermo Scientific Finnpette F2)
Thermomixer (Eppendorf, catalog number: 5355)
Thermal cycler S1000 (Bio-Rad, catalog number: 1852196)
Incubator (Thermo Scientific, catalog number: 51029323)
Shaker (GFL, catalog number: 3015)
Tabletop microcentrifuge (Eppendorf 5424)
Vortex (Scientific Industries, model: Vortex Genie 2)
Spectrophotometer (Spark, Tecan)
Incubator (Thermo Scientific Heraeus, BBD 6220)
Incubator settings: 37 °C, 5 % CO2 , 95 % relative humidity
Centrifuge (Eppendorf, 5810, equipped with aerosol-tight buckets located in a BSL2 laboratory)
Fully-equipped biosafety level 2 laboratory authorized for work with virus particles (BSL 2)
IQue Screener Plus [Intellicyt, equipped with plate reader, blue (488 nm) and red (640 nm) laser]
Software
Geneious Prime 2022.0.2
Forecyt standard Edition 7.0 (R2) (7.0.7035)
Microsoft Excel
R studio version: 1.3.1056
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Proietti, L., Manhart, G., Heyes, E., Troester, S. and Grebien, F. (2022). Arrayed CRISPR/Cas9 Screening for the Functional Validation of Cancer Genetic Dependencies. Bio-protocol 12(24): e4577. DOI: 10.21769/BioProtoc.4577.
分类
癌症生物学 > 通用技术 > 分子生物学技术
生物信息学与计算生物学
分子生物学 > DNA > DNA 克隆
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link