- EN - English
- CN - 中文
Profiling of Single-cell-type-specific MicroRNAs in Arabidopsis Roots by Immunoprecipitation of Root Cell-layer-specific GFP-AGO1
通过根细胞层特异性 GFP-AGO1 的免疫沉淀分析拟南芥根中单细胞类型特异性 MicroRNAs
发布: 2022年12月20日第12卷第24期 DOI: 10.21769/BioProtoc.4575 浏览次数: 2192
评审: Anonymous reviewer(s)
Abstract
MicroRNAs (miRNA) are small (21–24 nt) non-coding RNAs involved in many biological processes in both plants and animals. The biogenesis of plant miRNAs starts with the transcription ofMIRNA (MIR) genes by RNA polymerase II; then, the primary miRNA transcripts are cleaved by Dicer-like proteins into mature miRNAs, which are then loaded into Argonaute (AGO) proteins to form the effector complex, the miRNA-induced silencing complex (miRISC). In Arabidopsis, some MIR genes are expressed in a tissue-specific manner; however, the spatial patterns of MIR gene expression may not be the same as the spatial distribution of miRISCs due to the non-cell autonomous nature of some miRNAs, making it challenging to characterize the spatial profiles of miRNAs. A previous study utilized protoplasting of green fluorescent protein (GFP) marker transgenic lines followed by fluorescence-activated cell sorting (FACS) to isolate cell-type-specific small RNAs. However, the invasiveness of this approach during the protoplasting and cell sorting may stimulate the expression of stress-related miRNAs. To non-invasively profile cell-type-specific miRNAs, we generated transgenic lines in which root cell layer-specific promoters drive the expression of AGO1 and performed immunoprecipitation to non-invasively isolate cell-layer-specific miRISCs. In this protocol, we provide a detailed description of immunoprecipitation of root cell layer-specific GFP-AGO1 using EN7::GFP-AGO1 and ACL5::GFP-AGO1 transgenic plants, followed by small RNA sequencing to profile single-cell-type-specific miRNAs. This protocol is also suitable to profile cell-type-specific miRISCs in other tissues or organs in plants, such as flowers or leaves.
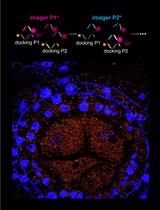
Graphical abstract

Background
MicroRNAs (MiRNAs) play an important role in many cellular growth and development processes. A fantastic feature of miRNAs is the non-cell autonomous action, in which miRNA moves from cell to cell or over long distances (Melnyk et al., 2011), serving as a signal molecule that functions locally or systemically. Previous studies have shown that the long distance or systemic movement of miRNA is mainly through the phloem, while cell-to-cell movement is through plasmodesmata (Melnyk et al., 2011). Mobile miRNAs, such as miR394 and miR165/6, have been shown to serve as morphogens, which shape the cell pattern formation during development (Breakfield et al., 2012; Carlsbecker et al., 2010; Knauer et al., 2013). Root cell layer-specific promoter-driven expression of AGO1, the core protein in the miRNA-induced silencing complex (miRISC), shows that AGO1 is cell autonomous (Brosnan et al., 2019; Fan et al., 2022). Microtubules have been shown to specifically regulate the association of miRNA165/6 with AGO1 in the cytoplasm and play a vital role in miRNA cell-to-cell movement (Fan et al., 2022). Therefore, characterization of miRNA distribution at a high spatial resolution is critical for understanding their functions in gene regulation. Here, we describe a protocol to profile cell-type-specific miRNA by immunoprecipitation of root cell layer-specific green fluorescent protein (GFP)-AGO1, followed by small RNAseq, that provides a new tool to deepen our understanding of the regulation of miRNA functions.
Materials and Reagents
RNase-free pipette tips [Genesee Scientific, catalog numbers: 23-130R (10 µL), 23-150R (200 µL), 23-165R (1,250 µL)]
1.5 mL centrifuge tubes (Genesee Scientific, catalog number: 22-282)
50 mL centrifuge tubes (Genesee Scientific, catalog number: 21-108)
15 mL centrifuge tubes (Genesee Scientific, catalog number: 21-103)
Petri dishes (Genesee Scientific, catalog number: 32-106)
Nylon mesh (mesh opening sizes 100 µm) (NITEX, catalog number: NTX100-098)
Liquid nitrogen
Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific, catalog number: F-530XL)
In-Fusion HD cloning plus (Takara, catalog number: 638911)
Murashige and Skoog medium (MS) (PhytoTech Lab, catalog number: M524)
IGEPAL CA-630 (MilliporeSigma, catalog number: I8896)
GFP-Trap magnetic agarose (ChromoTek, gtd-20)
Sodium dodecyl sulfate (SDS) (MilliporeSigma, catalog number: L3771-500G)
β-mercaptoethanol (MilliporeSigma, M3148-250ML)
Glycerol (Thermo Fisher Scientific, catalog number: J16374.K2)
Glycogen RNA grade (Thermo Fisher Scientific, catalog number: R0551)
NEBNext multiplex small RNA library prep set for Illumina (New England Biolabs, catalog number: E7300)
10 bp O'RangeRuler DNA ladder (Thermo Fisher Scientific, catalog number: SM1313)
Sodium chloride (NaCl) (Thermo Fisher Scientific, catalog number: AAJ2161836)
DEPC-treated water (Thermo Fisher Scientific, catalog number: 4387937)
Trizma base (MilliporeSigma, catalog number: T1503)
TRI reagent (MRC, catalog number: TR118)
Clorox germicidal bleach (Thermo Fisher Scientific, catalog number: Essendant CLO30966EA)
Triton X-100 solution (MilliporeSigma, catalog number: 93443)
Bromophenol blue (MilliporeSigma, catalog number: B0126)
cOmplete, EDTA-free protease inhibitor cocktail (MilliporeSigma, catalog number: 11836170001)
GFP antibody (MilliporeSigma, catalog number: 11814460001)
Chloroform (Thermo Fisher Scientific, catalog number: 423550040)
Isopropanol (Thermo Fisher Scientific, catalog number: 423835000)
6× DNA loading dye (Thermo Fisher Scientific, catalog number: R0611)
Ethidium bromide (Thermo Fisher Scientific, catalog number: J67270.14)
Sodium acetate (Thermo Fisher Scientific, catalog number: A13184.30)
Seed sterilization solution (see Recipes)
2× protein SDS loading buffer (see Recipes)
IP lysis buffer (see Recipes)
Washing buffer (see Recipes)
5× TBE buffer (see Recipes)
12% polyacrylamide gel (see Recipes)
8% SDS-PAGE resolving gel (see Recipes)
SDS-PAGE stacking gel (see Recipes)
3 M sodium acetate (pH 5.2) (see Recipes)
Equipment
Plant growth chamber (Percival, model: CU36L5)
Microcentrifuge (Eppendorf, model: 5424R)
High-speed centrifuge (Beckman Counter, model: Avanti J-E Series)
DynaMag-2 magnet (Invitrogen, catalog number: 12321D)
H5600 revolver tube mixer (VWR, catalog number: H5600-15)
Thermocyclers (Bio-Rad, catalog number: 1851148)
Blade
Thermomixer C (Eppendorf, catalog number: 5382000015)
Spatula
Mortar and pestle
Mini-PROTEAN tetra vertical electrophoresis cell (Bio-Rad, catalog number: 1658001FC)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Fan, L., Gao, B. and Chen, X. (2022). Profiling of Single-cell-type-specific MicroRNAs in Arabidopsis Roots by Immunoprecipitation of Root Cell-layer-specific GFP-AGO1. Bio-protocol 12(24): e4575. DOI: 10.21769/BioProtoc.4575.
分类
植物科学 > 植物分子生物学 > RNA > RNA 检测
植物科学 > 植物细胞生物学 > 细胞间通讯
分子生物学 > RNA > miRNA 干扰
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link