- EN - English
- CN - 中文
Efficient Generation of Genome-wide Libraries for Protein–ligand Screens Using Gibson Assembly
利用Gibson组装技术高效生成蛋白质配体筛选全基因组文库
发布: 2022年11月20日第12卷第22期 DOI: 10.21769/BioProtoc.4558 浏览次数: 3428
评审: Kristin L. ShinglerEmmanuel Orta-ZavalzaAnonymous reviewer(s)
Abstract
Genome-wide screens using yeast or phage displays are powerful tools for identifying protein–ligand interactions, including drug or vaccine targets, ligand receptors, or protein–protein interactions. However, assembling libraries for genome-wide screens can be challenging and often requires unbiased cloning of 105–107 DNA fragments for a complete representation of a eukaryote genome. A sub-optimal genomic library can miss key genomic sequences and thus result in biased screens. Here, we describe an efficient method to generate genome-wide libraries for yeast surface display using Gibson assembly. The protocol entails genome fragmentation, ligation of adapters, library cloning using Gibson assembly, library transformation, library DNA recovery, and a streamlined Oxford nanopore library sequencing procedure that covers the length of the cloned DNA fragments. We also describe a computational pipeline to analyze the library coverage of the genome and predict the proportion of expressed proteins. The method allows seamless library transfer among multiple vectors and can be easily adapted to any expression system.
Keywords: Genome-wide library (全基因组文库)Background
Protein–ligand interactions are essential for nearly all cellular processes, including gene expression regulation, pathogen–host interactions, and the identification of drug or vaccine targets. Defining interacting partners can be challenging and usually relies on in vitro biochemical approaches or genetic manipulation of the organism of interest. For many non-model organisms, genetic manipulation tools are still scarce or of laborious application, thus limiting the approaches available for discovering protein–ligand partners or protein interaction networks.
Genome-wide libraries are powerful tools for exploratory studies in protein–ligand interactions (Bharucha and Kumar, 2007; Bidlingmaier and Liu, 2011). Once incorporated into an expression system, libraries can encode all possible polypeptides of an organism without bias associated with its life cycle stage or culture conditions. Generating a eukaryote genome-wide library requires cloning approximately 105–107 different DNA fragments. Hence, the method must be robust to ensure that all possible coding sequences are included and in frame with the proteins of the expression system. The most common methods for generating genomic libraries involve DNA fragmentation by enzymatic digestion or sonication, followed by cloning the DNA fragments into restriction enzyme–digested or T/A-based plasmid vectors. Adapters or barcode sequences are often added to the DNA fragments to help with library amplification and sequencing.
There are several expression systems for interaction screenings. Baker's yeast—Saccharomyces cerevisiae—has been widely used as an expression system due to its versatility for genetic manipulation, high levels of protein expression, inducible gene expression, and the presence of eukaryotic post-translational modifications (Boder and Wittrup, 1997; Bidlingmaier and Liu, 2011). Yeast surface display (YSD) is one of the most successful screening systems for discovering protein–ligand interactions (Gibson et al., 2009; Bidlingmaier and Liu, 2011; Gu et al., 2014; Danecek et al., 2021). In this system, library proteins are expressed in frame with proteins of an endogenous surface anchoring system (e.g., Aga1p-Aga2p) and then displayed in approximately 105 copies of a single protein per cell (Inoue et al., 1990). The yeast population can express a whole genomic library and is usually screened using fluorescence or magnetic-activated cell sorting to enrich yeast cells that react with the ligands. The library DNAs from the sorted yeasts are then sequenced to identify the coding sequences for the ligand–interacting partners. Hence, the YSD methodology coupled with a strategy to enrich or isolate the positive interactions can lead to the discovery of protein–ligand partners while overcoming the limitations of genetic manipulation of non-model organisms.
We present here an efficient method to construct genome-wide libraries. In our approach, the genomic DNA is fragmented to the average gene size, end-repaired, and A-tailed for the ligation of adapter sequences for PCR amplification (Figure 1). The fragments are PCR-amplified with a pair of primers that hybridizes with the adapters and contains a ~20 bp overlapping sequence to the receptor vector for Gibson assembly. The Gibson assembly entails an isothermal reaction to join DNA molecules using a ~20 bp overlapping sequence between DNA fragments (e.g., library fragments and plasmid vector). This reaction includes a 5' exonuclease that generates 3’ overhangs in the joining molecules. After DNA molecules annealing, a DNA polymerase fills existent gaps in the annealed strands, and a DNA ligase seals strand nicks completing the process (Kieke et al., 1997). Using the Gibson method, the amplicons originating from adapter-ligated genomic fragments are joined to a linearized vector in a highly efficient one-step library assembly. Our protocol was designed to facilitate library transfer between expression systems and streamline library sequencing by Oxford nanopore technology to speed up the screening process. The nanopore sequencing technology has the advantage of generating sequences covering the entire cloned fragment; however, the protocol can be adapted to other sequencing methods. Our protocol details the library assembly, optimized library sequencing, and a computational pipeline to analyze the library coverage of the genome and evaluate library diversity and completeness. We also include a computational tool that predicts the library polypeptides expressed in frame with proteins of the expression system.
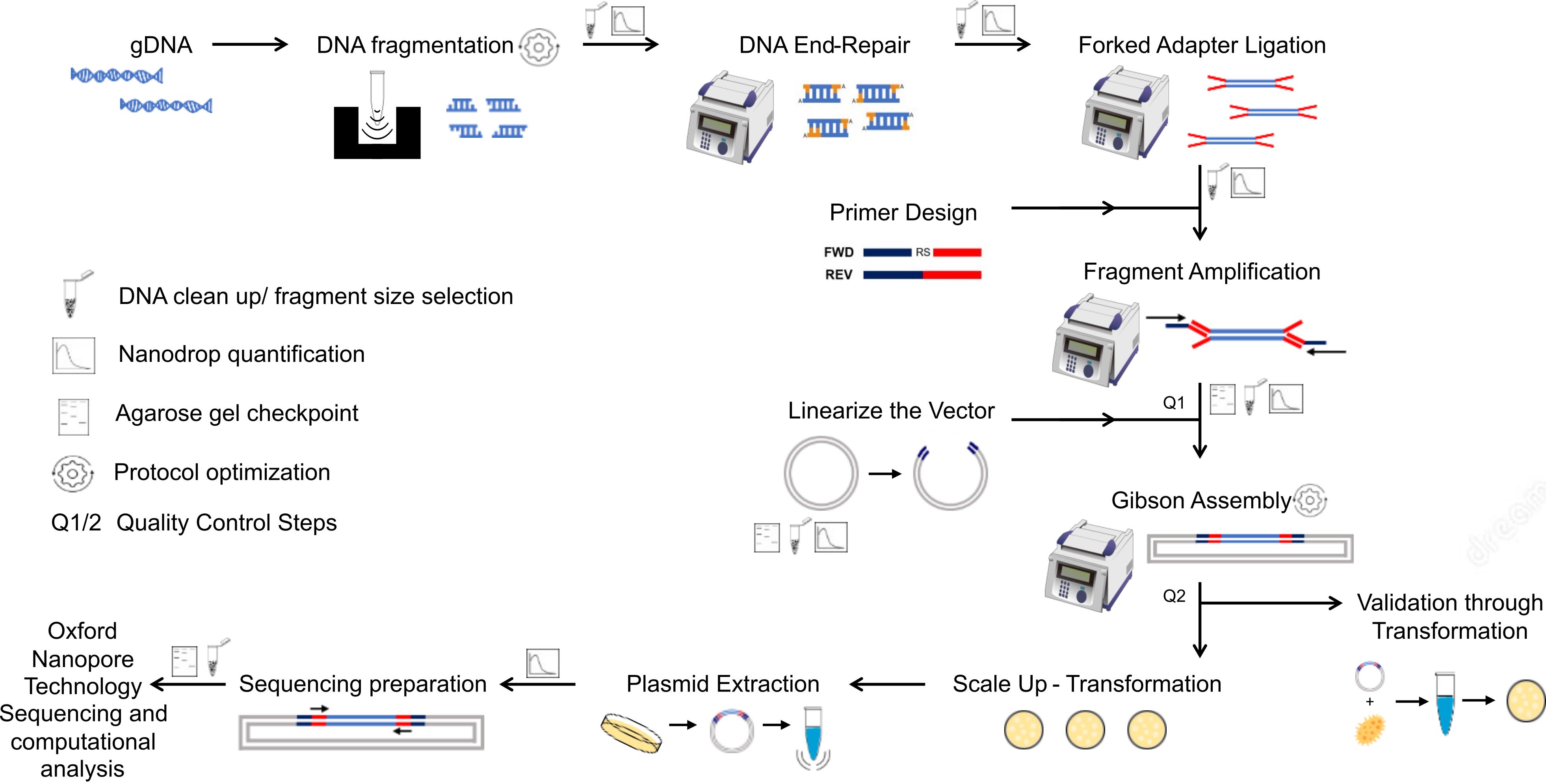
Figure 1. Diagram of genome-wide library preparation protocol and quality control steps
Materials and Reagents
Serological pipet, 10 mL (Drummond, catalog number: 6 000 010)
Labcon Eclipse refill universal-fit pipette tips with TubeGuard 0.1–10 µL (Labcon Eclipse, catalog number: 1036-260)
VWR refill pipette tips 100/200 µL (VWR, catalog number: 89079-476)
VWR refill pipette tips 1,000 µL (VWR, catalog number: 89079-470)
15 mL Centrifuge Tube, Bulk, Sterile, 25/pk, 500/cs (Montreal Biotech, catalog number: MBI601052C)
50 mL Centrifuge Tube, Bulk, Sterile, 25/pk, 500/cs (Montreal Biotech, catalog number: MBI602052C)
150 mm × 15 mm tissue culture-treated dishes (Fisher Scientific, catalog number: FB0875714)
Microtube-50 AFA fiber screw-cap vials (Covaris, catalog number: 520166)
Syringe Filter PES 0.22 µm 30 mm diameter, sterilized by Gamma irradiation (Ultident, catalog number: 229747)
NEB® 5-alpha F'Iq Competent Escherichia coli (High Efficiency) (New England Biolabs, catalog number: C2992H)
Mag-Bind® TotalPure NGS beads (Omega Bio-Tek, catalog number: M1378-01)
NEBNext® end-repair module (New England Biolabs, catalog number: E6050S)
Monarch® DNA elution buffer (New England Biolabs, catalog number: T1016L)
Ligation sequencing kit (Oxford Nanopore Technologies, catalog number: SQK-LSK110)
Native barcoding expansion 1-12 (Oxford Nanopore Technologies, catalog number: EXP-PBC001)
Blunt/TA ligase master mix (New England Biolabs, catalog number: M0367S)
NEBNext FFPE DNA repair mix (New England Biolabs, catalog number: M6630S)
NEBNext Quick Ligation Module – 20 rxs (New England Biolabs, catalog number: E6056S)
Taq DNA polymerase with ThermoPol buffer (New England Biolabs, catalog number: M0267S)
dNTPs mixture 10 mM (BioBasic, catalog number: DD0056)
Agarose (Bishop Canada Inc., catalog number: AGA002.250)
NEBuilder HiFi DNA assembly master mix (New England Biolabs, catalog number: E2621S)
Tryptone (BioBasic, catalog number: TG217(G211))
Yeast extract (BioBasic, catalog number: G0961)
Agar A (Biobasic, catalog number: FB0010)
Sodium chloride (Fisher Scientific, catalog number: S271-1)
Sodium Hydroxide (white pellets) (Fisher Scientific, catalog number: BP359-212)
Hydrochloric acid (Fisher Scientific, catalog number: A144-212)
Boric Acid (Omnipure, catalog number: 2710)
Ethylenediaminetetraacetic acid (EDTA) (Sigma Aldrich, catalog number: E9884-100G)
Manganesium chloride Hexahydrate (Fisher Scientific, BP214-500)
Calcium chloride dihydrate (Sigma Aldrich, catalog number: 223506-500G)
PIPES, Sesquisodium Sodium Salt (Fisher Scientific, catalog number: BP304-100)
Potasium hydroxide pellets (Fisher Scientific, catalog number: P250-500)
Trizma Base (AMRESCO, catalog number: 0497-5KG)
Ampicillin, trihydrate (BioBasic, catalog number: AB0064)
pUC-19 plasmid (NEB, catalog number: N3041S)
Covaris AFA-grade water (800 mL) (D-mark Biosciences, catalog number: Cov-520101)
Lambda DNA/Hind III plus marker (BioBasic, catalog number: M105R-2)
Ecostain (BioBasic, catalog number: DT81413)
Cell scraper (Cole-Parmer, catalog number: UZ01959-14)
EZ-10 spin column plasmid DNA miniprep kit (BioBasic, catalog number: BS414)
NucleoBond® Xtra maxi plus EF (Takara, catalog number: 740426.10)
pYD1 plasmid (Addgene, catalog number: 73447)
Restriction enzymes:
Bam HI (New England Biolabs, catalog number: R0136S)
Hind III (New England Biolabs, catalog number: R0104S)
Xho I (New England Biolabs, catalog number: R0146S)
NEBufferTM r3.1 (New England Biolabs, catalog number: B6003S)
Flow cell (R9.4.1) (Oxford Nanopore Technologies, catalog number: FLO-MIN106D)
Luria-Bertani (LB) medium with agar and ampicillin (see Recipes)
Ampicillin stock solution (100 mg/mL) (50 mL) (see Recipes)
Tris-Borate-EDTA buffer (10 L) (see Recipes)
10× TBE stock solution (1 L) (see Recipes)
SOB medium (1 L) (see Recipes)
Inoue solution (1 L) (see Recipes)
0.5 M PIPES buffer (100 mL) (see Recipes)
70% ethanol (see Recipes)
Equipment
Pipettes 0.2–2 µL (Gilson, catalog number: FA10001M), 2–20 µL (Gilson, catalog number: FA10003M), 20–200 µL (Gilson, catalog number: FA10005M), and 100–1,000 µL (Gilson, catalog number: FA10006M)
Portable Pipet-Aid® XP pipette controller (Drummond, catalog number: 4-000-101).
Focused-ultrasonicator (Covaris, M220, catalog number: 500295)
12-position magnetic rack for 1.5 mL tubes (Promega, catalog number: Z5342)
T100 thermal cycler (Bio-Rad, catalog number: 1861096)
Isotemp 215 water bath (Fisher, catalog number: 15-462-15)
Avanti® J-E centrifuge, 50 Hz, 200 V, 24 A (Beckman Coulter, catalog number: 369005)
Microcentrifuge 5418/5418R (Eppendorf, catalog number: EP5401000137)
Variable speed rocking platform shaker (VWR, catalog number: 75832-308)
NAPCO analog display automatic CO2 water-jacketed incubator (Napco, catalog number: 102219021)
Innova® 44 incubator shaker (New Brunswick, catalog number: M1282-0000)
Water system ultrapure (Millipore Synergy, catalog number: C9202)
Electrophoresis unit: Thermo Scientific Power Supply 400 mA 300 V (Fisher, catalog number: S65533Q)
NanoDrop ND-1000 (Thermo Fisher, catalog number: 2353-30-0010)
ChemiDocMP imaging system (BioRad, catalog number: 12003154)
Oxford nanopore MinION Mk1C sequencer (Oxford Nanopore Technologies, catalog number: MIN-101C)
Software
Minimap2 (Lemos Duarte et al., 2021) (https://github.com/lh3/minimap2)
Samtools (Li, 2018) (https://www.htslib.org/)
DeepTools (Liao et al., 2014) (https://deeptools.readthedocs.io/en/develop/index.html)
Libframe (This work) (https://github.com/cestari-lab/Libframe-tool)
Subread (Pointer et al., 2014) (http://subread.sourceforge.net/featureCounts.html)
Circlize (Ramirez et al., 2016) (https://jokergoo.github.io/circlize/)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Sternlieb, T., Loock, M., Gao, M. and Cestari, I. (2022). Efficient Generation of Genome-wide Libraries for Protein–ligand Screens Using Gibson Assembly. Bio-protocol 12(22): e4558. DOI: 10.21769/BioProtoc.4558.
分类
系统生物学 > 相互作用组 >
微生物学 > 异源表达系统 > 酿酒酵母
药物发现 > 药物设计
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link