- EN - English
- CN - 中文
Staphylococcus aureus 30S Ribosomal Subunit Purification and Its Biochemical and Cryo-EM Analysis
金黄色酿脓葡萄球菌中30S核糖体亚基的纯化及其生化特性和冷冻电镜分析
发布: 2022年10月20日第12卷第20期 DOI: 10.21769/BioProtoc.4532 浏览次数: 2811
评审: Alba BlesaKristin L. ShinglerNathan Rhys James
Abstract
The ribosome is a complex cellular machinery whose solved structure allowed for an incredible leap in structural biology research. Different ions bind to the ribosome, stabilizing inter-subunit interfaces and structurally linking rRNAs, proteins, and ligands. Besides cations such as K+ and Mg2+, polyamines are known to stabilize the folding of RNA and overall structure. The bacterial ribosome is composed of a small (30S) subunit containing the decoding center and a large (50S) subunit devoted to peptide bond formation. We have previously shown that the small ribosomal subunit of Staphylococcus aureus is sensitive to changes in ionic conditions and polyamines concentration. In particular, its decoding center, where mRNA codons and tRNA anticodons interact, is prone to structural deformations in the absence of spermidine. Here, we report a detailed protocol for the purification of the intact and functional 30S, achieved through specific ionic conditions and the addition of spermidine. Using this protocol, we obtained the cryo-electron microscopy (cryo-EM) structure of the 30S–mRNA complex from S. aureus at 3.6 Å resolution. The 30S–mRNA complex formation was verified by a toeprinting assay. In this article, we also include a description of toeprinting and cryo-EM protocols. The described protocols can be further used to study the process of translation regulation.
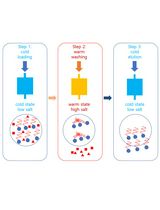
Graphical abstract:
Background
Protein synthesis is performed by the ribosome, which is composed of two dynamic subunits. These are made by RNAs and proteins that interact with several ligands and adopt several conformations during the translation process (Frank et al., 2007; Agirrezabala et al., 2008; Zhang et al., 2009). Structural and functional studies have revealed that ions impact the stability and dynamics of the ribosome (Nierhaus, 2014; Akanuma, 2021). Optimal ions and polyamine concentrations are necessary for maintaining both structural integrity and intrinsic flexibility. The small subunit (30S), responsible for the translation initiation process and the mRNA decoding, is particularly structurally dynamic and, thus, more challenging for structural studies. This subunit is composed of two main structural domains, the head and the body, which undergo continuous rearrangement (swiveling and nodding, respectively), heavily influencing the position of the mRNA in the decoding channel (Fahnestock et al., 1977; Hussain et al., 2016; Belinite et al., 2018). For this reason, specific ionic conditions have been used to stabilize the position of the mRNA for structural studies. Ribosome structure and dynamics are especially affected by the presence of cations, notably Mg2+, Zn2+, K+, NH4+, and polyamines (Gordon and Lipmann 1967; Weiss et al., 1973; Stahli and Noll 1977; Kim et al., 2007; Wohlgemuth et al., 2010; Nierhaus, 2014; Rozov et al., 2019). For instance, recent studies have demonstrated that different magnesium concentrations drastically affect the conformation of the 30S, and high temperatures help induce some conformational changes in Escherichia coli (Jahagirdar et al., 2020). In addition, spermidine has been shown to stabilize the active conformation of Staphylococcus aureus 30S by interacting with rRNA (Belinite et al., 2021); indeed, the correct folding of the decoding center at the top of the helix 44 (h44) in the 30S body can be achieved by maintaining a certain concentration of spermidine along the 30S purification process (Belinite et al., 2021). Spermidine and spermine are polyamines, organic polycations present in all cell types (Cohen and Lichtenstein, 1960; Stevens et al., 1969; Weiss and Morris, 1970; Cohen, 1971; Hardy and Turnock, 1971; Turnock and Birch, 1973), including in S. aureus (Herbst et al., 1958; Hamana and Satake 1995; Rosenthal and Dubin, 1962). These have been shown to associate with the ribosome (Cohen and Lichtenstein, 1960) and contribute to both the efficiency and fidelity of protein synthesis (Echandi and Algranati, 1975; Igarashi et al., 1980; Igarashi and Kashiwagi, 2018; McMurry and Algranati, 2018). They are known to stimulate in vitro translation (Hershko et al., 1961; Martin and Ames, 1962; Takeda, 1969; Igarashi et al., 1973; Konecki et al., 1975; Hetrick et al., 2010), lessening the Mg2+ requirement (Rheinberger and Nierhaus, 1987; Bartetzko and Nierhaus, 1988).
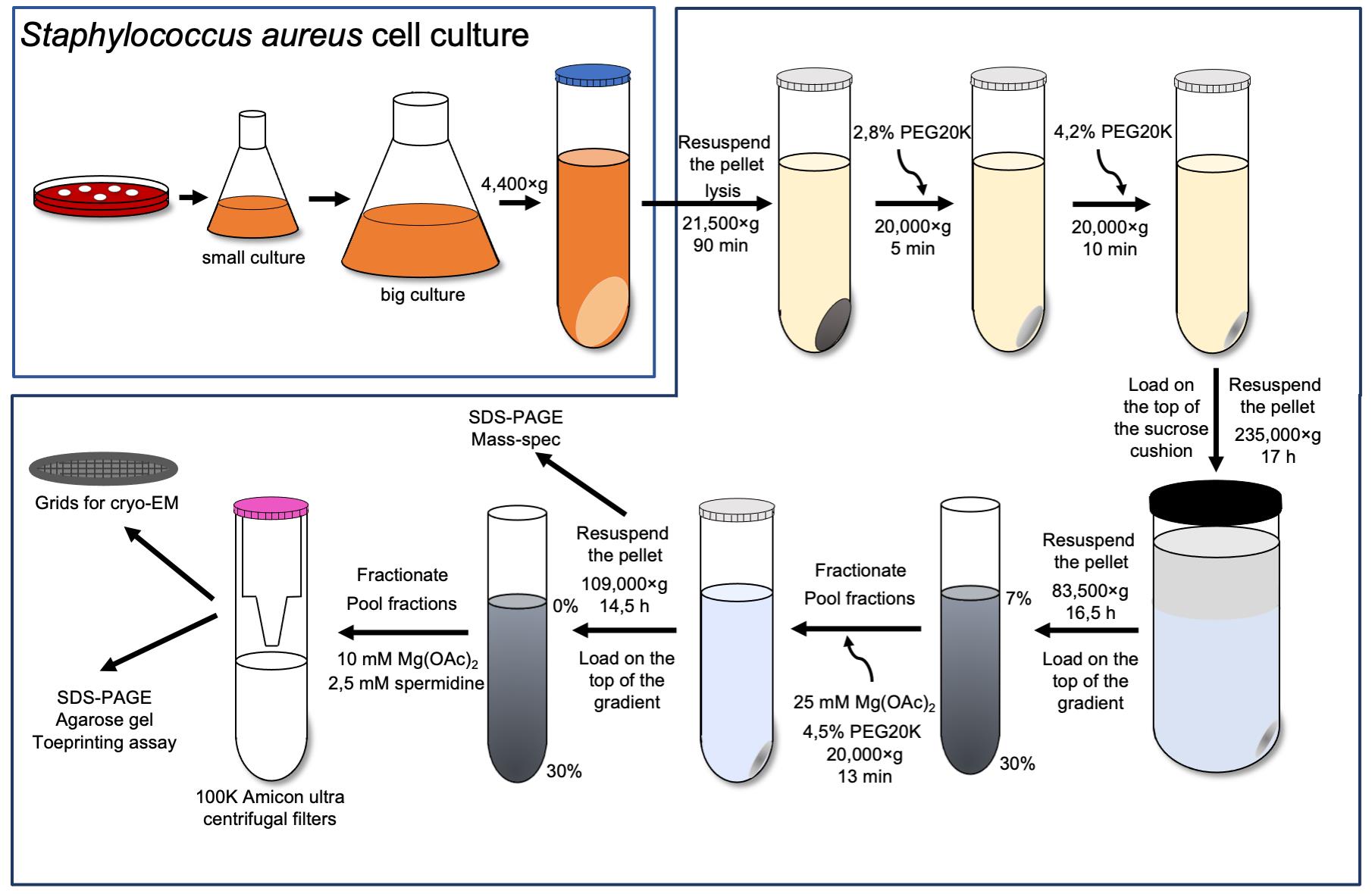
In this study, we describe in detail the protocols for the efficient 30S purification from S. aureus and the complex preparation with a natural S. aureus mRNA, spa mRNA, which encodes the virulence factor protein A. The proper conformation of the h44, full accommodation of the mRNA in the channel, and reduced flexibility of certain parts of the 30S were achieved by maintaining spermidine at a proper concentration throughout the whole process, starting from the 30S subunit isolation. The 30S subunit was obtained by separating the 70S ribosome under dissociative conditions and analyzed for high-molecular-weight contaminants and rRNA integrity. Activity of the isolated 30S was tested using a toeprinting assay (Fechter et al., 2009), while its structure was obtained by cryo-electron microscopy (cryo-EM). This technique has also been used to test different buffer conditions, thanks to the development of methods to rapidly get multiple structures (Cressey and Callaway, 2017; Nakane et al., 2020). The final S. aureus 30S–mRNA structure was obtained with focused refinement of the head and body at 3.6 and 3.4 Å resolution, respectively. Our previous work demonstrated the requirement of spermidine for the rRNA stabilization and its use in the cryo-EM studies (Belinite et al., 2021). Here, we show that the 30S subunits are stable in these conditions, confirming that spermidine not only contributes to their structural integrity, but also assures their required flexibility. Hence, protocols described in this article can be used for the future investigations of S. aureus functional complexes.
Materials and Reagents
Cells
Staphylococcus aureus RN6390 strain (Khusainov et al., 2016)
70S ribosome purification
Polycarbonate 50 mL centrifuge tubes (NalgeneTM, catalog number: 3117-0500)
Open-top thinwall ultra-clear tube, 38.5 mL (Beckman Coulter, catalog number: 344058)
Blood agar plates (BDTM Columbia agar with 5% sheep blood, catalog number: PA-254005.06)
Brain–heart infusion broth (BHI, Sigma-Aldrich, catalog number: 53286)
Lysostaphin (Sigma-Aldrich, catalog number: L7386)
DNase I (Roche, catalog number: 4716728001)
Protease inhibitor cocktail without ethylenediaminetetraacetic acid (EDTA, Thermo ScientificTM, catalog number: 78429)
30% PEG 20000 (Hampton Research, HR2-609)
Buffer A (see Recipes)
Buffer B (see Recipes)
5× buffer E (see Recipes)
Dissociation buffer (see Recipes)
30S subunit purification
Open-top thinwall ultra-clear tube, 13.2 mL (Beckman Coulter, catalog number: 344059)
Amicon ultra-15 centrifugal filter unit (Merck Millipore, catalog number: UFC910008)
Liquid nitrogen
30S storage buffer (see Recipes)
mRNA purification
RNaseZap (Thermo ScientificTM, catalog number: AM9780)
NucleoSpin Plasmid, mini kit for plasmid DNA (Macherey-Nagel, catalog number: 740588.50)
NucleoSpin Gel and PCR Clean-up, mini kit for gel extraction and PCR clean-up (Macherey-Nagel, catalog number: 740609.50)
BamHI enzyme (NEB, catalog number: R0136S)
CutSmart® buffer (NEB, catalog number: B7204S)
MEGAscript kit with DNase turbo (Ambion, catalog number: AM1333)
ROTI® phenol/chloroform/isoamyl alcohol, 250 mL, pH 7.5–8.0 (Carl Roth, catalog number: A156.1)
ROTI® Aqua-P/C/I, 250 mL, pH 4.5–5.0 (Carl Roth, catalog number: X985.1)
3 M sodium acetate
Buffer ELU (see Recipes)
30S and mRNA quality analysis
PageRulerTM protein ladder (Thermo ScientificTM, catalog numbers: 26616 and 26619)
InstantBlue® coomassie protein stain (Abcam, catalog number: ab119211)
ROTI® aqua-phenol, 250 mL, pH 4.5–5.0 (Carl Roth, catalog number: A980.1)
Chloroform (Thermo ScientificTM, catalog number: AC158210010)
Agarose (SeaKemTM, catalog number: 11590717)
1 M potassium thiocyanate (Sigma, catalog number: P3011)
Ethidium bromide (Thermo ScientificTM, catalog number: 15585011)
100% formamide (Sigma, catalog number: F9037)
RiboRuler low-range RNA ladder (Thermo ScientificTM, catalog number: SM1831)
4% stacking gel (see Recipes)
15% resolving gel (see Recipes)
4× blue buffer (see Recipes)
10× running buffer (see Recipes)
6% polyacrylamide gel (see Recipes)
10× TBE (see Recipes)
Toeprinting assay
10 μM Toe primer (5′ CCTGCAGGTCGACTC 3′)
10× PNK buffer (Thermo ScientificTM, catalog number: EK 0032)
10 U/μL T4 PNK (Thermo ScientificTM, catalog number: EK 0032)
[γ-32P]-ATP (370 MBq/mL, 10 mCi/mL, 185 MBq)
Micro bio-spin chromatography columns (Bio-Rad, catalog number: 732-6200)
20 U/μL AMV reverse transcriptase, isolated from Avian Myeloblastosis Virus (QBIOgene, catalog number: EMAMV200)
10× AMV buffer (QBIOgene, catalog number: EMAMV200)
10 mM dATP, dCTP, dGTP, and dTTP (Roche, catalog numbers: 11051440001, 11051458001, 11051466001, and 11051482001)
0.3 M Na acetate (Sigma, catalog number: S2889)
Phenol/chloroform, pH 4.5–5.0 (Carl Roth, catalog number: X985.1)
Chloroform/isoamyl alcohol (Carl Roth, catalog number: X984.1)
100% cold ethanol
80% cold ethanol
8 M urea (Sigma, catalog number: U5128)
0.1% bromophenol blue (Sigma, catalog number: B0126)
0.1% xylene cyanol (Sigma, catalog number: X4126)
100 µM ddATP, ddGTP, ddCTP, and ddTTP (Roche, catalog number: 3732738001)
3 M KOH (Sigma, catalog number: 221473)
1 M tris-HCl, pH 8.0 (Sigma, catalog number: T1503)
10% SDS (Bio-Rad, catalog number: 1610416)
Acetic acid (Sigma, catalog number: A6283)
10× toeprinting buffer (see Recipes)
30S–mRNA complex preparation for cryo-EM
Liquid ethane
Liquid nitrogen
Quantifoil R2/2300-mesh holey carbon grids
Equipment
70S ribosome purification
2 L flasks
Microbiological incubator (Thermo ScientificTM, catalog number: 50125882 or equivalent)
Cell culture shaker at 30 °C
Class II biosafety cabinet
Floor centrifuges (Beckman, Avant J20 XP and/or Avanti J-E or equivalent)
JA-25.50 rotor (Beckman, catalog number: 363055)
JLA-16.250 rotor (Beckman, catalog number: 363930)
Ultracentrifuge (Beckman, OPTIMA XE90 or equivalent)
Type 45 Ti rotor (Beckman, catalog number: 339160)
SW28 rotor (Beckman, catalog number: 342204)
Gradient MasterTM base unit (Biocomp, catalog number: B108-2)
Gradient MasterTM device (BioComp Instruments, Fredericton, Canada)
Fraction collector (Bio-Rad, model: 2110)
NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA or equivalent)
30S subunit purification
Ultracentrifuge (Beckman, OPTIMA XE90 or equivalent)
SW41 Ti swinging-bucket rotor (Beckman, catalog number: 331362)
Floor centrifuges (Beckman, Avanti J-E or equivalent)
JLA-16.250 rotor (Beckman, catalog number: 363930)
mRNA purification
Cell culture shaker at 37 °C
Benchtop centrifuge (Eppendorf MiniSpin® Plus personal microcentrifuge with 12-place rotor or equivalent)
Eppendorf thermomixer R, dry block heating and cooling shaker or equivalent
30S and mRNA quality analysis
Mini-PROTEAN® Tetra Handcast systems (Bio-Rad or equivalent)
PowerPacTM basic power supply (Bio-Rad or equivalent)
Wide mini-sub cell GT cell (Bio-Rad or equivalent)
GelDoc Go Gel imaging system (Bio-Rad)
Toeprinting assay
Benchtop centrifuge (Eppendorf MiniSpin® Plus personal microcentrifuge with 12-place rotor or equivalent)
Phosphor imaging or autoradiography (Typhoon or equivalent)
30S–mRNA complex preparation cryo-EM data acquisition
Thermo Scientific Vitrobot Mark IV
Talos Arctica equipped with Falcon III direct electron detector (Thermo Fisher)
Microscope control PC with installed EPU software
Software
MotionCor2 (https://emcore.ucsf.edu/ucsf-software/)
Gctf (https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/)
Relion 3.0 (https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page/)
ResMap (http://resmap.sourceforge.net/)
Chimera (https://www.cgl.ucsf.edu/chimera/)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Belinite, M., Khusainov, I. and Marzi, S. (2022). Staphylococcus aureus 30S Ribosomal Subunit Purification and Its Biochemical and Cryo-EM Analysis. Bio-protocol 12(20): e4532. DOI: 10.21769/BioProtoc.4532.
分类
生物化学 > 蛋白质 > 分离和纯化
生物物理学 > 显微技术 > 低温显微镜技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link