- EN - English
- CN - 中文
A New Tool for the Flexible Genetic Manipulation of Geobacillus kaustophilus
一种对嗜热土杆菌进行灵活遗传操作的新工具
(*contributed equally to this work) 发布: 2022年09月05日第12卷第17期 DOI: 10.21769/BioProtoc.4502 浏览次数: 1902
评审: Alessandro DidonnaAksiniya AsenovaAnonymous reviewer(s)
Abstract
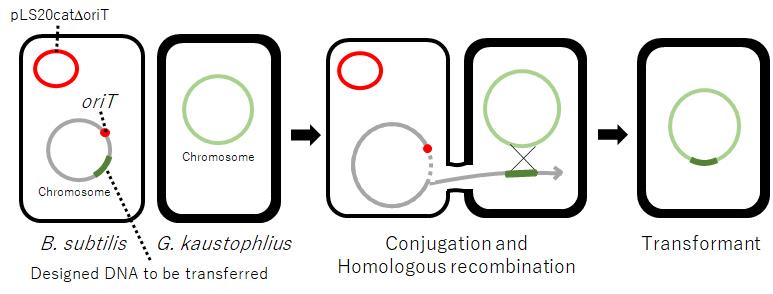
Geobacillus kaustophilus, a thermophilic Gram-positive bacterium, is an attractive host for the development of high-temperature bioprocesses. However, its reluctance against genetic manipulation by standard methodologies hampers its exploitation. Here, we describe a simple methodology in which an artificial DNA segment on the chromosome of Bacillus subtilis can be transferred via pLS20-mediated conjugation resulting in subsequent integration in the genome of G. kaustophilus. Therefore, we have developed a transformation strategy to design an artificial DNA segment on the chromosome of B. subtilis and introduce it into G. kaustophilus. The artificial DNA segment can be freely designed by taking advantage of the plasticity of the B. subtilis genome and combined with the simplicity of pLS20 conjugation transfer. This transformation strategy would adapt to various Gram-positive bacteria other than G. kaustophilus.
Graphical abstract:
Background
Geobacillus kaustophilus HTA426 is a thermophilic Gram-positive bacterium isolated from sediments of the Mariana Trench (Nazina et al., 2001; Takami et al., 2004). It grows at temperatures ranging from 48 to 74 °C (optimal temperature of 60 °C) and is a promising chassis for high-temperature bioprocessing with the advantages of preventing contamination by mesophilic bacteria and lowering the cost of controlling fermentation heat. However, this bacterium is difficult to transform by natural competence or electroporation, and an approach based on a conjugation system for Gram-negative bacteria has been developed (Suzuki and Yoshida, 2012). This method has the disadvantage of not being able to select transformants that acquired mutations with adverse effects on growth. Therefore, better techniques for manipulating the genome of G. kaustophilus are needed.
Conjugation is one of the three main mechanisms for horizontal gene transfer in bacteria. Conjugative elements, which often are located on plasmids, encode all the proteins required for their transfer from a donor cell to a recipient cell. Conjugative elements encode adhesins or organelles facilitating interaction with recipient cells. In almost all conjugative systems only a single strand of the DNA (ssDNA) is transferred. Generation of the ssDNA is done by a relaxase, often assisted by one or two auxiliary proteins, that introduces a site- and strand-specific nick in a region of the plasmid named origin of transfer (oriT). After nicking, the relaxase remains covalently attached to the 5′ end of the DNA, and the generated 3′ end is used as a primer for DNA synthesis resulting in a rolling-circle mode of replication, thereby generating the ssDNA molecule that is destined for transfer into the recipient cell. Conjugative elements encode a sophisticated transferosome (type IV secretion system) connecting the donor and recipient cells. The relaxosome complex, which is composed of a number of proteins including the relaxase, is recruited to the cytosolic side of the transferosome in the donor cell, after which it guides the transfer of the ssDNA into the recipient cell (Christie et al., 2014; Christie, 2016; Waksman, 2019).
pLS20 is a 65 kb conjugative plasmid isolated from Bacillus subtilis var. natto (Tanaka and Koshikawa, 1977); it can transfer itself among various Bacillus species (Koehler and Thorne, 1987). Contrary to most conjugative systems that require prolonged interactions between donor and recipient cells on solid medium, efficient conjugation of pLS20 is achieved by mixing donor and recipient cells in liquid medium (Tanaka and Koshikawa, 1977; Singh et al., 2013; Miyano et al., 2018). Genes involved in pLS20 conjugation (genes 28–74) form a large operon controlled by a strong promoter named Pc. Regulation of pLS20 conjugation genes has been studied in detail (for a review, see Meijer et al., 2021). Gene 27 encodes RcopLS20, which is the master regulator of the conjugation operon. In its default state, conjugation is suppressed due to binding of RcopLS20 to its two operator sites within the Pc region, strictly repressing the Pc promoter. Gene 25 encodes an antirepressor named RappLS20, which activates conjugation by relieving repression from RcopLS20 and forming a Rap/Rco complex. Gene 26 encodes a quorum sensing peptide, Phr*pLS20, that regulates the activity of RappLS20. phrpLS20 synthesizes a small pre-proprotein. After being secreted, PhrpLS20 is proteolytically cleaved, generating the mature Phr*pLS20 peptide corresponding to its 5 C-terminal residues. When taken up by donor cells, Phr*pLS20 inactivates RappLS20 by changing its conformation upon binding to it, returning the system to its default repressed state (Singh et al., 2013). Ectopic overproduction of RappLS20 increased the efficiency of conjugation by weakening the effect of Phr*pLS20 (Singh et al., 2013).
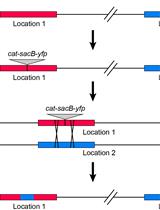
A derivative of pLS20 lacking oriTpLS20, pLS20catΔoriT, is defective in conjugation (Miyano et al., 2018). However, we have recently shown that pLS20catΔoriT can mediate the transfer of large regions of the bacterial chromosome between B. subtilis cells when a copy of oriTpLS20 is present on the chromosome in the donor cell (Miyano et al., 2018). Inspired by this, we have developed a strategy to transform G. kaustophilus that allows for flexible manipulation of its chromosome (Mori et al., 2022). Here, we present the protocol to transform G. kaustophilus by pLS20-mediated conjugative transfer of a chromosomal DNA of B. subtilis designed to function in G. kaustophilus.
Materials and Reagents
Note: All reagents can be stored at room temperature unless otherwise specified.
Construction of the donor strain
B. subtilis 168 (Mori et al., 2022)
B. subtilis YNB211 (Mori et al., 2022)
G. kaustophilus MK244 (Mori et al., 2022)
pUCG18T (Mori et al., 2022)
LB medium (2% agar) (BD Difco, catalog number: 240230)
10% SDS (Nacalai Tesque, catalog number: 30562-04)
Lysozyme (Wako, catalog number: 129-06723)
Phenol/Chloroform/Isoamyl alcohol (25:24:1) (Wako, catalog number: 311-90151), store at 4 °C
70% Ethanol
99.5% Ethanol
KOD -Plus- Neo polymerase (Toyobo, catalog number: KOD-401), store at -20 °C
Agarose for electrophoresis gel (ME, Nacalai Tesque, catalog number: 01133-24)
In silico Molecular Cloning software for designing PCR primers (In Silico Biology, Inc., catalog number: IMCGF01)
Primer pairs used for the PCR reaction of five fragments
Fragment A1:
aprE-U-f1 ccggtacttgccaccacatcataac
aprE-U-r cagtaacctcatcaagccaagctacctctcgctatttccgtagagactcg
Fragment A2:
aprE-D-f2 gacagaggaattagatacattcgcgttaatcaacgtacaagcagctgcac
aprE-D-r ggccgagcagtattcgaatgtcaag
Fragment B1:
degA-U-f gtagcttggcttgatgaggttactg
degA-U-r2 catcggtcataaaatccgtatccttggttactttcatcgctcatcattc
Fragment B2:
degA-D-f2 ctgcaaggcgattaagttgggtaacagtgaaatcgtaaggatgtgagcag
degA-D-r cgcgaatgtatctaattcctctgtc
Fragment C1:
kan-f aaggatacggattttatgaccgatg
kan-r2 gttacccaacttaatcgccttgcag
Primers used for the recombinant PCR reactions that ligate the five fragments.
aprE-U-f3-nested caccgagctcatagcttgtcgcgatcacctcatcc
aprE-D-r2-nested tgctttcgctgattacaacattggtgacgctgcct
DNA purification kit (Wizard® SV Gel and PCR Clean-Up System, Promega, catalog number: A9281)
7.5 mg/mL Kanamycin sulfate stock solution in water (Wako, catalog number: 113-00343)
Solution A (see Recipes)
Solution B (see Recipes)
100× Trace element (see Recipes)
TKE buffer (see Recipes)
50× TAE buffer (see Recipes)
C-1 and C-2 medium for transformation (see Recipes)
Conjugation and selection of transconjugant (transformant)
7.5 mg/mL Kanamycin sulfate stock solution in water (Wako, catalog number: 113-00343)
10 mg/mL Chloramphenicol stock solution in 70% ethanol (Wako, catalog number: 030-19452)
100 mM IPTG stock solution in water (Nacalai Tesque, catalog number: 367-93-1)
LB medium (see Recipes)
LBMSM medium (see Recipes)
Southern blotting analysis
Primers used for amplification of a template DNA for synthesizing RNA probe
Km-probe-f taatacgactcactatagggtatggctctcttggtcgtc
Km-probe-r tctgattccacctgagatgc
Agarose for electrophoresis gel (SP, Nacalai Tesque, catalog number: 01163-76)
DNA Molecular Weight Marker II, DIG-labeled (Roche, catalog number: 11218590910)
DNA purification kit (Wizard® SV Gel and PCR Clean-Up System, Promega, catalog number: A9281)
T7 RNA polymerase (Roche, catalog number: 10881767001), store at -20 °C
DIG RNA Labeling Mix, 10× conc. (Roche, catalog number: 11277073910), store at -20 °C
Restriction enzymes
Cla I (Takara, catalog number: 1034A), store at -20 °C
Sac II (Takara, catalog number: 1079A), store at -20 °C
DIG Easy Hyb (Roche, catalog number: 11603558001), store at 4 °C
Amersham Hybond-N+ (GE Healthcare, catalog number: RPN82B)
Hybridization bag (Hybridization Bags Hybri-Bag Hard, COSMO BIO, catalog number: S-1001)
Blocking Reagent (Roche, catalog number: 11096176001)
Anti-Digoxigenin-AP, Fab fragment (Roche, catalog number: 11093274910), store at 4 °C
CSPD, ready-to-use (Roche, number: 11755633001), store at 4 °C
20× SSC buffer (see Recipes)
Maleic acid buffer (see Recipes)
Denaturing solution (see Recipes)
Neutralizing solution (see Recipes)
High-stringency wash solution (see Recipes)
Low-stringency wash solution (see Recipes)
Detection buffer (see Recipes)
Equipment
Construction of the donor strain
Thermal cycler (for example, Takara Bio, Thermal Cycler Dice Touch TP350, or equivalent)
Centrifuge (for example, TOMY, MX-307, or equivalent)
Spectrophotometer (for example, GE Healthcare, NanoVue Plus, or equivalent)
Agarose gel electrophoresis apparatus (for example, Takara Bio, Mupid®-2plus, or equivalent)
Incubator (for example, PHCbi, MIR-H163, or equivalent)
Conjugation and selection of transconjugant (transformant)
Two incubator shakers, one set at 37 °C and the other at 60 °C (TAITEC, Bioshaker BR-43FM-MR, or equivalent)
Centrifuge (TOMY, MX-307, or equivalent)
Two incubators, one is set at 37 °C and the other 60 °C (PHCbi, MIR-H163, or equivalents)
Spectrophotometer (SHIMADZU, UV-1800, or equivalent)
300 mL baffled Erlenmeyer flask
Southern blotting (or: Southern blot analysis)
Thermal cycler (Takara Bio, Thermal Cycler Dice Touch TP350, or equivalent)
Spectrophotometer (GE Healthcare, NanoVue Plus, or equivalent)
Agarose gel electrophoresis apparatus (Bio-Rad, Sub-Cell GT Cell, or equivalent)
Transfer apparatus (Bio Craft, Vacuum Transfer BS-31, or equivalent)
UV crosslinker (UVP, CL-1000, or equivalent)
Shaker (Bio Craft, Labo Shaker BC-730, or equivalent)
Chemiluminescence imager (Bio-Rad, ChemiDoc XRS+, or equivalent)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Amatsu, R., Mori, K., Ishikawa, S., Meijer, W. J. J. and Yoshida, K. (2022). A New Tool for the Flexible Genetic Manipulation of Geobacillus kaustophilus. Bio-protocol 12(17): e4502. DOI: 10.21769/BioProtoc.4502.
分类
微生物学 > 微生物遗传学 > 转化
分子生物学 > DNA > 转染
分子生物学 > DNA > DNA 重组
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link