- EN - English
- CN - 中文
CRISPR/Cas9-mediated Gene Knockout Followed by Negative Selection Leads to a Complete TCR Depletion in orthoCAR19 T Cells
CRISPR/Cas9 介导的基因敲除后负选择导致原CAR19 T 细胞中的 TCR 完全耗尽
发布: 2022年08月05日第12卷第15期 DOI: 10.21769/BioProtoc.4485 浏览次数: 3894
评审: Alak MannaNavnita DuttaAnonymous reviewer(s)
Abstract
Genome-editing technologies, especially CRISPR (clustered regularly interspaced short palindrome repeats)/Cas9 (CRISPR-associated protein 9), endows researchers the ability to make efficient, simple, and precise genomic DNA changes in many eukaryotic cell types. CRISPR/Cas9-mediated efficient gene knockout holds huge potential to improve the efficacy and safety of chimeric antigen receptor (CAR) T cell-based immunotherapies. Here, we describe an optimized approach for a complete loss of endogenous T cell receptor (TCR) protein expression, by CRISPR/Cas9-mediated TCR α constant (TRAC) and TCR β constant (TRBC) gene knockout, followed by subsequent CD3 negative selection in engineered human orthoCAR19 T cells. We believe this method can be expanded beyond CAR T cell application, and target other cell surface receptors.
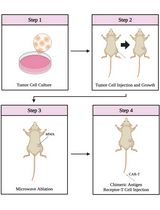
Graphical abstract:

Schematic overview of the two-step process of endogenous TCR depletion in engineered human orthoCAR19 T cells using (1) CRISPR/Cas9-mediated gene knockout followed by (2) CD3 negative selection.
Background
CRISPR-Cas9 technology enables gene knockouts (KO) and complete loss of protein translation in human cells, especially in primary cells (Seki and Rutz, 2018). In recently published research, we showed that combining an orthogonal human IL-2 and IL-2Rβ (ortho-hIL-2/ortho-hIL-2Rβ) system with CAR19 T cell therapy markedly enhanced the efficacy of the CAR T cell antitumor activity, in a well-established preclinical model of acute lymphoblastic leukemia (ALL) (Zhang et al., 2021). However, the unexpected deaths observed with high-dose ortho-hIL-2 led us to investigate mechanisms of toxicity. To explore whether T cell-mediated toxicity is related to acute xenogeneic graft-versus-host disease (GVHD) mediated through the T cell receptor (TCR), we knocked out the endogenous TCR of CAR T cells co-expressing ortho-hIL-2Rβ, using a CRISPR-Cas9 gene KO approach. We use guide RNAs (gRNA) targeting the TCR α constant (TRAC) and TCR β constant (TRBC) loci, followed by CD3 negative selection, resulting in TCR negative orthoCAR19 T cells.
Gene silencing is a powerful approach to study gene function and to discern molecular mechanisms underlying complex human diseases. Previous studies have utilized RNA interference (RNAi), small interfering RNA (siRNA), or short hairpin RNA (shRNA) technologies to effectively knock down genes of interest (Berns et al., 2004; O’Keefe, 2013). However, these knockdown approaches do not result in complete loss of gene/protein expression or KO and can often result in off-target effects (Jackson et al., 2006).
The CRISPR-Cas9 genome editing system consists of two components: a “guide” RNA (gRNA) and a Cas9. The Cas9 protein is an endonuclease that uses gRNA to form base pairs with DNA target sequences, enabling Cas9 to introduce a site-specific double-stranded break in the DNA. Through RNA-directed Cas9 nucleases, the CRISPR-Cas9 system can modify DNA with greater precision than existing technologies, such as transcription activator-like endonuclease (TALEN), and zinc-finger nuclease (ZFN) (Conant et al., 2022). While both TALEN and ZFN are artificial structures formed by joining restriction endonucleases with DNA-binding protein domains, CRISPR-Cas9 technology takes advantage of the antiviral defense mechanism of prokaryotes to perform genome editing at the targeted DNA sequence (Khan, 2019). Recent work done by Cui et al. revealed that both TALEN and ZFN inevitably generated massive off-target effects when targeting human papillomavirus 16 (HPV16), while the CRISPR-Cas9 system was shown to be more efficient and specific (Cui et al., 2021). Therefore, CRISPR-Cas9 technology shows better efficiency, feasibility, and practicality in multi-role clinical applications, as compared to TALEN and ZFN. Inference of CRISPR Edits (ICE) was used to analyze CRISPR editing data in this study. ICE is a free and easy-to-use software tool that offers fast and reliable analysis of CRISPR experiments, highly comparable to that of next-generation sequencing (NGS) data. CRISPR editing can be assessed and analyzed by uploading the Sanger sequencing data and specifying a guide sequence(s). A knockout score will be generated, which represents the proportion of cells that have either a frameshift or insertion-deletion mutation (indels). This score is a useful means to quantify the number of contributing indels that are likely to result in a functional KO of the targeted gene.
Our results showed that the CRISPR-Cas9 system is easy to operate, has a short cycle time, low cost, higher accuracy, and is more controllable in targeting TCR α and TCR β genes together than existing technologies such as TALEN and ZFN. The combination of CRISPR/Cas9 with CD3 negative selection leads to a complete TCR depletion in orthoCAR19 T cells.
Materials and Reagents
Tissue culture plates, flasks, and tubes
24 well cell tissue culture plates (Corning, catalog number: 3524)
T25 cell culture flasks (Thermo Fisher, catalog number: 156367)
T75 cell culture flasks (Thermo Fisher, catalog number: 156499)
15 mL centrifuge tubes (Corning, catalog number: 352095)
50 mL centrifuge tubes (Corning, catalog number: 430829)
Phosphate Buffered Saline solution (PBS) (Corning, catalog number: 21-031-CM)
RPMI 1640 medium (Gibco, catalog number: 11875085)
Bovine Calf Serum (Sigma, catalog number: 12306C-500ml)
Penicillin/Streptomycin (Thermo Fisher, catalog number: 10378016)
L-glutamine (Thermo Fisher, catalog number: A2916801)
Recombinant human Interleukin-2 (IL2) (R&D System, catalog number: 202-IL-050)
APC anti-human TCR α/β antibody (Biolegend, catalog number: 306718)
DynabeadsTM CD3 (Thermo Fisher, catalog number:11151D)
TrueCutTM Cas9 Protein v2 (Invitrogen, catalog number: A36497)
EasySet Magnet (Thermo Fisher, catalog number: NC9284020)
DNeasy Blood & Tissue Kit (Qiagen, catalog number: 69504)
Q5 Hot Start High-Fidelity DNA Polymerase Kit (BioLabs, catalog number: E0555S)
Trypan Blue Solution, 0.4% (Thermo Fisher, catalog number: 15250061)
QIAquick PCR Purification Kit (Qiagen, catalog number: 28106)
4D-NucleofectorTM X Kit (Lonza, catalog number: V4XC-2032)
Cell culture media (see Recipes)
Wash buffer for cells (see Recipes)
gRNA (see Recipes)
Equipment
4D-NucleofectorTM X Unit (Lonza, catalog number: AAF-1002X)
Flow Cytometer LSRFortessa (BD Biosciences, catalog number: 647800L6)
EppendorfTM 5424 Microcentrifuges (Fisher, catalog number: 05-400-005)
Cell culture hood HeraSafe KS (Thermo Fisher, catalog number: 10110910)
CO2 incubator HeraCell VIOS 160i (Thermo Fisher, catalog number: 15381075)
VeritiProTM 96-well Thermal Cycler (Applied Biosystems, catalog number: A47394)
Software
Genewiz (Azenta Life Sciences, https://www.genewiz.com/en)
ICE analysis (Synthego, https://ice.synthego.com/#/)
CRISPOR (Tefor, http://crispor.tefor.net/)
FlowJo (Tree Star Inc., version 10.1)
Prism 9 for macOS V.9.3.1(350) (https://www.graphpad.com/scientific-software/prism/)
BioRender (https://biorender.com/)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhang, Q., Yang, J., Anand Manoharan, E. N. E., Yu, A. B. and Milone, M. C. (2022). CRISPR/Cas9-mediated Gene Knockout Followed by Negative Selection Leads to a Complete TCR Depletion in orthoCAR19 T Cells. Bio-protocol 12(15): e4485. DOI: 10.21769/BioProtoc.4485.
- Zhang, Q., Hresko, M. E., Picton, L. K., Su, L., Hollander, M. J., Nunez-Cruz, S., Zhang, Z., Assenmacher, C. A., Sockolosky, J. T., Garcia, K. C., et al. (2021). A human orthogonal IL-2 and IL-2Rβ system enhances CAR T cell expansion and antitumor activity in a murine model of leukemia. Sci Transl Med 13(625): eabg6986.
分类
癌症生物学 > 肿瘤免疫学 > 癌症治疗 > 细胞移植治疗
免疫学 > 免疫细胞功能 > 抗原特异反应
分子生物学 > DNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link