- EN - English
- CN - 中文
Preparation of a Single Cell Suspension from the Murine Iridocorneal Angle
从小鼠虹膜角膜角制备单细胞悬液
发布: 2022年05月20日第12卷第10期 DOI: 10.21769/BioProtoc.4426 浏览次数: 2824
评审: Pilar Villacampa AlcubierreCarolina TecuatlAnonymous reviewer(s)

相关实验方案
自动层次分析(ALAn):用于无偏见表征培养中哺乳动物上皮结构的图像分析工具
Christian Cammarota [...] Tara M. Finegan
2024年04月20日 4218 阅读
Abstract
Single cell RNA sequencing is a powerful tool that can be used to identify distinct cell types and transcriptomic differences within complex tissues. It has proven to be especially useful in tissues of the eye, where investigators have identified novel cell types within the retina, anterior chamber, and iridocorneal angle and explored transcriptomic contribution to disease phenotypes in age-related macular degeneration. However, to obtain high quality results, the technique requires isolation of healthy single cells from the tissue of interest, seeking complete tissue digestion while minimizing stress and transcriptomic changes in the isolated cells prior to library preparation. Here, we present a protocol developed in our laboratory for isolation of live single cells from the murine iridocorneal angle, which includes Schlemm’s canal and the trabecular meshwork, suitable for single cell RNA sequencing, flow cytometry, or other downstream analysis.
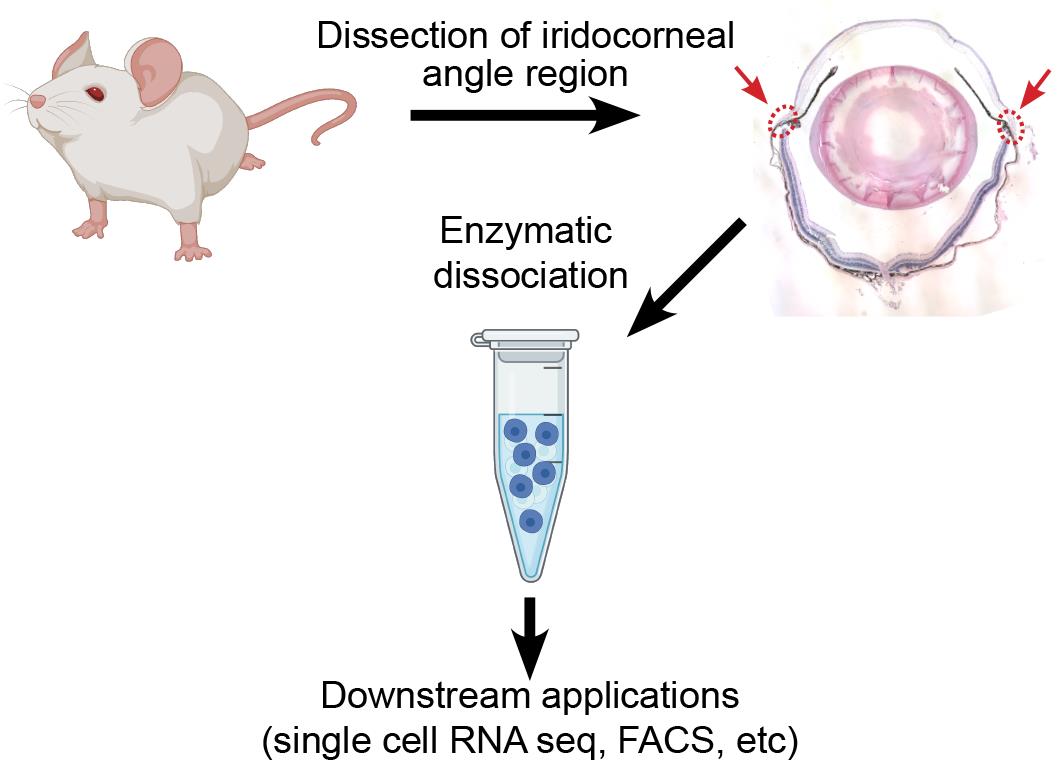
Graphical abstract:
Background
Glaucoma is a progressive neurodegenerative disease with no cure, affecting over 60 million individuals worldwide and leaving 8 million blind (Quigley and Broman, 2006). Current therapy is focused on reduction of intraocular pressure (IOP), the only treatable risk factor for disease progression. IOP is determined by the ratio between aqueous humor secretion by the ciliary body and drainage through tissues in the iridocorneal angle, including the trabecular meshwork and Schlemm’s canal, which comprise the conventional outflow pathway. Patients with glaucoma, including primary congenital glaucoma, exhibit defects in tissues of the conventional outflow pathway leading to decreased aqueous humor outflow and elevated IOP (Tektas and Lütjen-Drecoll, 2009; Braunger et al., 2015; Zagora et al., 2015). The importance of these tissues to glaucoma pathogenesis, and consequently as therapeutic targets, has focused intense research interest in understanding their physiology, molecular phenotypes, and the cell-cell interactions governing their function. However, due to the small sizes of the tissues involved and their location within the iridocorneal angle, relatively little is known about the transcriptomes or overall phenotypes of Schlemm’s canal or drainage channel endothelial cells, potential differences between inner and outer wall endothelial cells of Schlemm’s canal or cells of the trabecular meshwork.
Recently, single cell RNA sequencing has been used to identify cell types in the iridocorneal angle region of mice, humans, and other model species (van Zyl et al., 2020; Patel et al., 2020; Thomson et al., 2021). These studies have provided exciting hints about the cells making up the angle and identified potential new therapeutic targets, but the extreme rarity of Schlemm’s canal endothelial cells within published datasets [van Zyl et al. (2020) identified just 25 Schlemm’s canal endothelial cells after sequencing 13,833 cells (0.18%), while our group has identified 109 in a dataset of 19,232 (0.57%)] has emphasized the need for sequencing of more cells, in healthy and disease conditions, to better understand the full complexity of this important tissue.
In the present protocol, we optimized a straightforward method for dissecting and dissociating the murine iridocorneal angle to obtain a single cell suspension of healthy cells to be used for downstream applications such as single cell RNA sequencing or flow cytometry. Previous studies utilizing the full anterior segment have reported that corneal and ocular surface epithelial cells represent over 50% of total cells in the dataset (van Zyl et al., 2020). While glaucoma-relevant iridocorneal angle cell populations remain rare, by removing the majority of the cornea during dissection, our method substantially increases their yield, resulting in a ~200% increase in proportion of Schlemm’s canal endothelial cells and ~80% increase in trabecular meshwork cells within the sample. Depending on the tissue(s) of interest, this protocol could be combined with flow cytometry or antibody-conjugated bead-based methods to enrich the sample in relevant cell types prior to library preparation. We have also found that this procedure can be readily adapted to dissociate cells of the cornea or posterior eye with similar results and cell viability.
Materials and Reagents
Pipet tip micro 40 μm cell strainers (Bel-Art Flomi, catalog number: 136800040)
Standard 10 cm Petri dishes (one per sample)
Standard 20 mm tissue culture dishes (one per sample)
Standard 1.5 mL microcentrifuge tubes
Standard 1 mL pipet tips (ThermoFisher ART 2079 or similar)
Wide orifice 1 mL pipet tips (ThermoFisher ART 2079GPK or similar)
DMEM, dye free (ThermoFisher, Gibco, catalog number: 21063029)
Fetal bovine serum (ThermoFisher, Gibco, catalog number: 26140079 or similar)
0.25% Trypsin EDTA (ThermoFisher, Gibco, catalog number: 25200056)
10 μm Rock inhibitor Y-27632 (ATCC, catalog number: ACS-3030)
DNase 1 (Roche, catalog number: 04716728001)
Collagenase A (Roche, catalog number: 10103578001)
HBSS (ThermoFisher, Gibco, catalog number: 14175095)
Bovine Serum Albumen (Sigma-Aldrich, catalog number: 03117332001)
Acridine orange/propidium iodide live/dead staining kit compatible with your automated cell counter (i.e., Nexcelom CS2-0106)
Compatible disposable slides if needed for your automated cell counter (i.e., Nexcelom CHT4-PD100)
Digestion mix (see Recipes)
Resuspension solution (see Recipes)
25 mg/mL Collagenase A stock solution (see Recipes)
10 mM Y27632 stock solution (see Recipes)
Note: All reagents stored per manufacturer’s recommendations and used before labeled expiry dates.
Equipment
Shaking hot block (e.g., Eppendorf Thermomixer 5384000020)
Dissecting Microscope (we prefer a microscope with 6.3×–40× variable zoom optics for this method)
Refrigerated microcentrifuge capable of 350 × g (e.g., Eppendorf 5425R)
Automated cell counter (Nexcelom Cellometer Auto2000 or similar with compatible fluorescent live/dead staining kit)
Vannas scissors, curved (Fine Science Tools, catalog number: 15019-10 or similar)
Fine Bonn Scissors, straight or straight bladed Vannas scissors (Fine Science Tools, catalog number: 14084-08 or similar).
Fine forceps (Dumont #1, #5 or similar)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Thomson, B. R. and Quaggin, S. E. (2022). Preparation of a Single Cell Suspension from the Murine Iridocorneal Angle. Bio-protocol 12(10): e4426. DOI: 10.21769/BioProtoc.4426.
分类
发育生物学 > 细胞信号传导
细胞生物学 > 组织分析 > 组织成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link