- EN - English
- CN - 中文
A Highly Sensitive Method to Efficiently Profile the Histone Modifications of FFPE Samples
一种高效分析 FFPE 样品组蛋白修饰的高灵敏度方法
(*contributed equally to this work) 发布: 2022年05月20日第12卷第10期 DOI: 10.21769/BioProtoc.4418 浏览次数: 3181
评审: Alessandro DidonnaWenfei JiPierre-Damien DenechaudAnonymous reviewer(s)

相关实验方案
髓系肿瘤中用于生殖系 DNA 分离的皮肤成纤维细胞标准化培养:一种从床旁到实验室的实用方法
Parampreet Kour [...] Pulkit Rastogi
2025年10月05日 1411 阅读
Abstract
The majority of biopsies in both basic research and translational cancer studies are preserved in the format of archived formalin-fixed paraffin-embedded (FFPE) samples. Profiling histone modifications in archived FFPE tissues is critically important to understand gene regulation in human disease. The required input for current genome-wide histone modification profiling studies from FFPE samples is either 10–20 tissue sections or whole tissue blocks, which prevents better resolved analyses. Nevertheless, it is desirable to consume a minimal amount of FFPE tissue sections in the analysis as clinical tissue of interest are limited. Here, we present FFPE tissue with antibody-guided chromatin tagmentation with sequencing (FACT-seq), highly sensitive method to efficiently profile histone modifications in FFPE tissue by combining a novel fusion protein of hyperactive Tn5 transposase and protein A (T7-pA-Tn5) transposition and T7 in vitro transcription. FACT-seq generates high-quality chromatin profiles from different histone modifications with low number of FFPE nuclei. We showed a very small piece of FFPE tissue section containing ~4000 nuclei is sufficient to decode H3K27ac modifications with FACT-seq. In archived FFPE human colorectal and human glioblastoma cancer tissue, H3K27ac FACT-seq revealed disease specific super enhancers. In summary, FACT-seq allows researchers to decode histone modifications like H3K27ac and H3K27me3 in archival FFPE tissues with high sensitivity, thus allowing us to understand epigenetic regulation.
Graphical abstract:
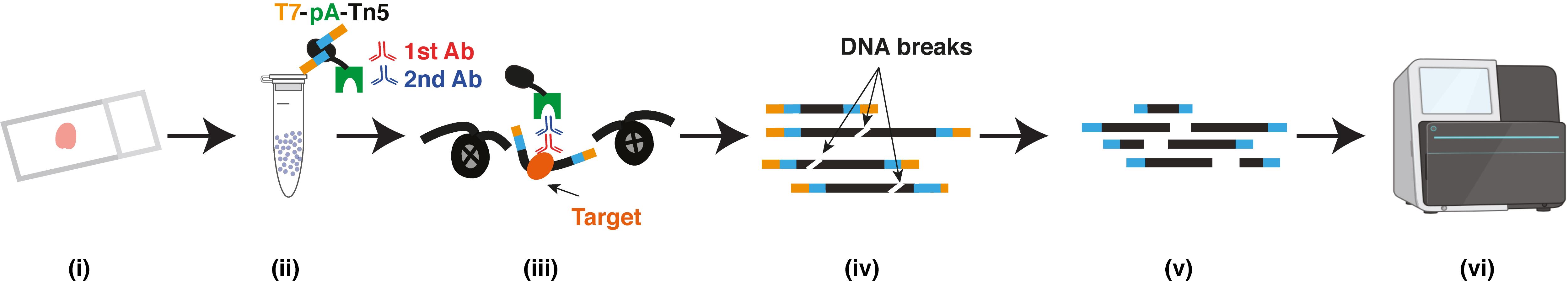
(i) FFPE tissue section; (ii) Isolated nuclei; (iii) Primary antibody, secondary antibody and T7-pA-Tn5 bind to targets; (iv) DNA purification; (v) In vitro transcription and sequencing library preparation; (vi) Sequencing
Background
Currently, Formalin-Fixed Paraffin-Embedding (FFPE) is a universal approach for biopsy specimen processing in basic research and translational studies (Fox et al., 1985; Wang et al., 2005; Haile et al., 2019). Furthermore, it has been reported that large numbers of FFPE specimens are archived worldwide each year (Waldron et al., 2012). More and more techniques have been developed to decipher genomic(Astolfi et al., 2015; Bolognesi et al., 2016), transcriptomic (Pennock et al., 2019), and proteomic (Becker et al., 2017) information in FFPE samples. Nevertheless, a method that can profile epigenetic regulation in FFPE samples with high sensitivity is lacking. Previously, researchers applied chromatin immunoprecipitation with sequencing (ChIP-seq) to archived FFPE tissue. And they have successfully developed pathology tissue chromatin immunoprecipitation (PAT-ChIP) (Fanelli et al., 2010, 2011), fixed-tissue chromatin immunoprecipitation sequencing (FiT-seq) (Cejas et al., 2016), and fixed-tissue ChIP-seq for H3K27 acetylation profiling (FiTAc-seq) (Font-Tello et al., 2020) making it possible to profile histone modifications in FFPE tissue. However, the required input for these technologies from FFPE samples is either 10–20 tissue sections or whole tissue blocks. A higher sample consumption prevents them from being used in a broader application since clinical samples are usually limited. In addition, sonication is included in these available methods, which may potentially introduce sequencing bias (Teytelman et al., 2009). Here, we present a new highly sensitive method, we recently developed, to efficiently profile the histone modifications from FFPE samples using antibody-guided chromatin tagmentation with sequencing (FACT-seq) (Zhao et al., 2021). FACT-seq uses a novel fusion protein (T7-Pa-Tn5) that combines hyperactive Tn5 transposase with protein A, utilizing T7 in vitro transcription to prepare a library, and profile histone modifications in a small amount of FFPE sample.
Materials and Reagents
50 mL Falcon tubes (Sarstedt, catalog number: 62.559.001)
21 G needles (BD Microlance, catalog number: ND432)
27 G needles (BD Microlance, catalog number: 302200)
1 mL syringes (BD Microlance, catalog number: 309628)
1.5 mL Low-bind tubes (Sarstedt, catalog number: 72.706.600)
0.5 mL Qubit tubes (Invitrogen, catalog number: Q32856)
30 μm Filters (Miltenyi Biotech MACS, catalog number: 130-098-458) (room temperature)
Xylene (Histolab, catalog number: 2070) (Room temperature)
Ethanol (VWR BDH Chemicals, catalog number: VWRC20816.552) (room temperature)
Collagenase (Sigma-Aldrich, catalog number: C9263)(-20°C)
Hyaluronidase (Merck Millipore, catalog number: HX0154) (-20°C)
Ampicillin (Serva, catalog number: 69-52-3) (+4°C)
Sodium azide (Merck Millipore, catalog number: 822335) (room temperature)
BSA (Miltenyi Biotech MACS, catalog number: 130-091-376) (+4°C)
IGEPAL CA-630 (Sigma-Aldrich, catalog number: I3021) (+4°C)
1 M CaCl2 (Alfa Aesar, catalog number: J63122) (room temperature)
5 M NaCl (Thermo Fisher Scientific, Invitrogen, catalog number: AM9759) (room temperature)
1 M Tris-HCl pH 8.0 (Thermo Fisher Scientific, Invitrogen, catalog number: 15568-025)
1 M Tris-HCl pH 7.5 (Thermo Fisher Scientific, Invitrogen, catalog number: 15567-027)
1 M MgCl2 (Thermo Fisher Scientific, Invitrogen, catalog number: AM9530G) (room temperature)
FBS (Life Technologies, catalog number: 10108-105) (-20°C)
RNase A (Thermo Fisher Scientific, catalog number: EN0531) (-20°C)
DTT (Thermo Fisher Scientific, catalog number: 20291) (+4°C)
0.5 M EDTA (Thermo Fisher Scientific, Invitrogen, catalog number: AM9260G) (room temperature)
Dimethylformamide (Sigma-Aldrich, catalog number: D4551) (room temperature)
Glycerol (Sigma-Aldrich, catalog number: G9012) (Room temperature)
Sodium deoxycholate (Sigma-Aldrich, catalog number: D6750) (room temperature)
Triton X-100 (Sigma-Aldrich, catalog number: T8787) (room temperature)
Human genomic DNA (Promega, catalog number: G3041) (+4°C)
Qiagen miniElute PCR purification kit (Qiagen, catalog number: 28004)
6× Loading Dye (Thermo Fisher Scientific, catalog number: R0611) (-20°C)
HEPES (Sigma-Aldrich, catalog number: H3375)
Protease inhibitor (Sigma-Aldrich, catalog number: 11873580001)
Spermidine (Sigma-Aldrich, catalog number: S2626)
10% SDS (Thermo Fisher Scientific, Invitrogen, catalog number:1553-035)
Anti-H3K27ac antibody (Abcam, catalog number: ab4729)
Anti-H3K27me3 antibody (Cell Signalling Technology, catalog number: 9733S)
Anti-H3K4me1 antibody (Abcam, catalog number: ab176877)
Anti-H3K36me3 antibody (Abcam, catalog number: ab9050)
Mouse IgG1, kappa monoclonal (Abcam, catalog number: ab18443)
Guinea Pig anti-Rabbit IgG antibody (Antibodies-Online, catalog number: ABIN101961)
Proteinase K (Thermo Fisher Scientific, catalog number: EO0491)
Phenol (Thermo Fisher Scientific, catalog number: 17914)
Chloroform (Sigma-Aldrich, catalog number: C2432)
NEBNext high-fidelity 2× PCR master mix (New England Biolabs, catalog number: M0541S) (-20°C)
SPRIselect beads (Beckman Coulter, catalog number: B23317) (Room temperature)
T7 high yield RNA synthesis kit (New England Biolabs, catalog number: E2040S) (-20°C)
Zymo RNA purification kit (Zymo Research, catalog number: R1013) (room temperature)
SMART MMLV kit (TAKARA, catalog number: 639524) (-20°C)
RNAClean XP beads (Beckman Coulter, catalog number: A63987) (+4°C)
Zymo ChIP DNA clean and concentrator kit (Zymo Research, catalog number: D5205) (room temperature)
40% Acrylamide:bis-acrylamide (Invitrogen, catalog number: HC2040)
10% Ammonium persulfate (Invitrogen, catalog number: HC2005)
TEMED (Invitrogen, catalog number: HC2006)
Digitonin (Millipore, catalog number: 300410)
Nuclease-free water (Invitrogen, catalog number: AM9932)
KOH (Sigma-Aldrich, catalog number: 484016)
50 bp DNA ladder (ThermoFisher Scientific, catalog number: 10488099)
Agilent high sensitive DNA kit (Agilent, catalog number: 5067-4626)
Tn5 transposase [produced in local protein facility following previously description (Picelli et al., 2014)]
pA-Tn5 transposase [produced in local protein facility following previously description (Picelli et al., 2019)]
1× PBS (pH = 7.4) (Thermo Fisher Scientific, catalog number: 10010023)
SYBR gold (ThermoFisher Scientific, catalog number: S33102)
10× TBE Buffer (ThermoFisher Scientific, catalog number: B52)
Mouse kidney/liver FFPE tissue blocks (prepared in the lab)
Buffer used in isolation of single nuclei suspension from FFPE tissue (see Recipes)
Buffers used in Tn5 assembly and activity assay (see Recipes)
Buffers used in epitope retrieval (see Recipes)
Buffers used in antibody binding and tagmentation (see Recipes)
Buffer used in library preparation (see Recipes)
Equipment
Microtome (Leica, Histo Core MULTICUT semi-automated rotary microtome, catalog number:149MULTI0C1, 14051856372 or similar one)
Stereo Microscope (Zeiss stemi DV4 stereo microscope 8x-32x, catalog number: 435421-0000-000 or similar one)
ThermoMixer (Eppendorf, Thermo mixer F1.5, catalog number: 5385000016 or similar one)
Thermal Cycler (Applied Biosystems, Veriti, catalog number: 4375786 or similar thermal cycler)
NanoDrop (Thermo Fisher scientific, Nanodrop 2000c, catalog number: ND2000CLAPTOP or similar one)
Cell Counter (Life Technologies, Countess II, catalog number: AMQAF1000 or similar one)
Centrifuge (Eppendorf centrifuge 5415R, catalog number: EP5415R or similar one)
Rotator (Grant InstrumentsTM 360° Vertical Multi-function Rotator PTR 35, catalog number: 9.721028 or similar one)
Microscopy (Zeiss Imager.Z2, catalog number: 490016-0001-000 or similar one)
DynaMagTM-2 magnetic rack (Thermo Fisher Scientific, catalog number: 12321D or similar one)
Agilent DNA Bioanalyzer (Agilent, catalog number: 5067-4626)
Polyacrylamide gel tray (Thermo Fisher Scientific, catalog number: HC1000S)
Gel documentation system (BIO-RAD, catalog number: 1708195EDU)
Software
Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtmL)
Samtools (http://samtools.sourceforge.net/)
Deeptools (https://deeptools.readthedocs.io/en/develop/content/installation.htmL)
Picard tools (https://broadinstitute.github.io/picard/)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Zhao, L., Polavarapu, V. K., Yadav, R. P., Xing, P. and Chen, X. (2022). A Highly Sensitive Method to Efficiently Profile the Histone Modifications of FFPE Samples. Bio-protocol 12(10): e4418. DOI: 10.21769/BioProtoc.4418.
分类
癌症生物学 > 通用技术 > 分子生物学技术
医学
分子生物学 > DNA > 染色质可接近性
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link