- EN - English
- CN - 中文
Production and Crystallization of Nanobodies in Complex with the Receptor Binding Domain of the SARS-CoV-2 Spike Protein
与 SARS-CoV-2 刺突蛋白受体结合域复合的纳米抗体的产生和结晶
(*contributed equally to this work) 发布: 2022年05月05日第12卷第9期 DOI: 10.21769/BioProtoc.4406 浏览次数: 3702
评审: Joana Alexandra Costa ReisLuke A YatesSneha Ray

相关实验方案
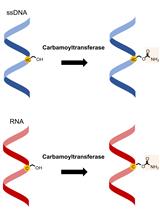
氨基甲酰转移酶测定: 5-羟甲基胞嘧啶 (5hmC) 到 5-氨基甲酰氧基甲基胞嘧啶 (5cmC) 的体外修饰
Weiwei Yang [...] Laurence Ettwiller
2022年09月05日 1982 阅读
Cell-Sonar:通过特定蛋白标志物表达变化追踪目标蛋白的简便低成本方法
Sabrina Brockmöller [...] Simone Rothmiller
2025年02月05日 1654 阅读
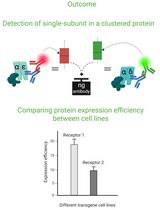
Cluster FLISA——用于比较不同细胞系蛋白表达效率及蛋白亚基聚集状态的方法
Sabrina Brockmöller and Lara Maria Molitor
2025年11月05日 1205 阅读
Abstract
The receptor binding domain (RBD) of the spike protein of SARS-CoV-2 binds angiotensin converting enzyme-2 (ACE-2) on the surface of epithelial cells, leading to fusion, and entry of the virus into the cell. This interaction can be blocked by the binding of llama-derived nanobodies (VHHs) to the RBD, leading to virus neutralisation. Structural analysis of VHH-RBD complexes by X-ray crystallography enables VHH epitopes to be precisely mapped, and the effect of variant mutations to be interpreted and predicted. Key to this is a protocol for the reproducible production and crystallization of the VHH-RBD complexes. Based on our experience, we describe a workflow for expressing and purifying the proteins, and the screening conditions for generating diffraction quality crystals of VHH-RBD complexes. Production and crystallization of protein complexes takes approximately twelve days, from construction of vectors to harvesting and freezing crystals for data collection.
Keywords: SARS CoV-2 (SARS-CoV-2)Background
Camelids (llamas, alpacas, and camels) produce a unique type of heavy chain only antibody that comprises variable domains (VHH) linked to Fc constant domains (CH2 and CH3). The VHHs can be produced as single domain antigen-binding proteins or nanobodies (Muyldermans, 2013), with wide applications in the life sciences (Pleiner et al., 2015; Jovcevska and Muyldermans, 2020). In response to the COVID-19 pandemic (Dhama et al., 2020), we and many other groups have developed VHH reagents that bind to the receptor binding domain (RBD) of the SARS-CoV-2 spike protein that block binding to ACE-2 at the cell surface, which is the primary interaction that leads to cell infection (Xiang et al., 2020; Hanke et al., 2020; Schoof et al., 2020; Koenig et al., 2021; Huo et al., 2020, 2021). Multimeric versions of these VHHs, either as trimers or IgG Fc fusions, have been shown to neutralise SARS-CoV-2 both in vitro and in animal models of COVID-19 (Huo et al., 2021; Nambulli et al., 2021). Structural analysis of VHH-RBD complexes has revealed two regions where epitopes are clustered, one at or close to the ACE-2 binding surface (cluster 2), and one at the opposite side of the RBD (cluster 1) (Tang et al., 2021). Information about the VHH binding sites has enabled the effect of mutations in the spike protein to be interpreted and predicted. In our experience, forming RBD complexes with two VHH that bind to orthogonal sites has been necessary for the crystallization of some VHH-RBD complexes (Huo et al., 2021).
VHHs are routinely produced in the E. coli strain WK6 as hexahistidine tagged proteins, using Isopropyl β-D-1-thiogalactopyranoside (ITPG) inducible promoters, and the addition of a secretion signal (e.g., pelB or ompA), to enable recovery of the VHH from the periplasm following induced expression. Primary purification uses immobilised metal affinity chromatography (IMAC) with gel filtration added as a polishing step. Alternatively, VHHs can be directly purified from lysed cells though, in our experience, periplasmic extraction by osmotic shock gives higher protein yields.
As a glycosylated protein, the production of the RBD of the SARS-CoV-2 spike protein requires expression in higher eukaryotic cells for which both insect and mammalian cells have been used. An appropriate signal sequence is added to the N-terminus to direct product to the cell media, and a hexahistidine tag added to the C-terminus for purification by IMAC. The N-glycosylation of the RBD (Asn331 and Asn343) introduces chemical heterogeneity, particularly if produced in mammalian cells, which generally inhibits crystallization (Chang et al., 2007). By growing cells in the presence of the mannosidase inhibitor kifunensine, N-glycosylation can be arrested at a high mannose state (GlcNAc2Man9 glycoforms). These can subsequently be trimmed back to single N-acetylglucosamine residues, by treatment with endoglycosidase F1 or H (Chang et al., 2007). Based on past experience (Nettleship et al., 2013), this is most efficiently carried out prior to purification of the VHH-RBD complex.
An issue for the purification of secreted glycoproteins from cell culture media by IMAC is that some media components displace the His-tagged protein-of-interest during the chromatography step, significantly reducing the yield. Here, we have used an automated method of affinity purification by IMAC and gel filtration, which involves loading the sample onto the Ni-NTA column in batches, with a column washing step between each batch (Nettleship et al., 2009).
We have reported the crystallization of a number of VHH-RBD complexes using commercially available screens, in 96-well format, and low sample volumes of 1–200 nL. Diffraction data were collected from the primary crystal hits using synchrotron X-rays, and structures solved by molecular replacement (Huo et al., 2020, 2021). Key to achieving good quality crystals was the preparation of the proteins. Thus, in this article, we describe our optimised workflow for producing VHH-RBD complexes and their crystallization for analysis by X-ray crystallography.
Materials and Reagents
Production of a receptor binding domain
Vector construction
pOPINTTG vector digested with KpnI/PmeI (Figure 1A)
Human codon optimised synthetic RBD gene (encoding amino acids 330–352) with 15-bp extensions overlapping with pOPINTTG in-fusion entry sites (lower case):
5’gcgtagctgaaaccggcCCGAATATCACAAATCTTTGTCCTTTCGGAGAAGTATTCAACGCAACTCGCTTCGCATCAGTATATGCCTGGAACCGCAAACGAATTTCTAATTGTGTCGCCGACTACTCTGTGCTTTACAACTCAGCATCATTTTCAACATTCAAATGCTATGGTGTCTCCCCTACGAAGTTGAACGATCTTTGTTTCACTAATGTCTACGCCGATTCCTTTGTTATTAGGGGGGACGAGGTTCGCCAGATCGCGCCGGGCCAAACGGGCAAGATAGCTGACTATAATTATAAGCTGCCAGACGACTTTACAGGCTGCGTTATCGCTTGGAATTCAAACAATCTTGATAGCAAAGTGGGTGGTAATTATAACTACCTCTACAGACTGTTTCGCAAGTCTAATCTGAAGCCTTTCGAGCGGGACATCTCTACTGAGATCTATCAAGCTGGTTCAACCCCCTGCAATGGCGTCGAGGGTTTTAACTGTTACTTCCCACTTCAGTCATACGGATTTCAACCAACTAATGGGGTTGGCTATCAGCCGTACCGCGTGGTCGTTCTTAGCTTTGAGCTGCTTCACGCCCCTGCAACGGTGTGCGGACCGAAAAAAAGTACAAAaaacatcaccatcac 3’
ClonExpress II One Step Cloning kit (Vazyme, catalog number: C113-02)
NucleoSpin 148® Gel and PCR Clean-up kit (MACHEREY-NAGEL, catalog number: 12303368)
Stellar competent cells (Takara Bio, catalog number: 636766)
LB medium (Sigma-Aldrich, catalog number: L3022)
S.O.C. (Super Optimal broth with Catabolite repression) recovery medium (ThermoFisher Scientific, catalog number: 15544034)
Ampicillin (Sigma-Aldrich, catalog number: A9393)
LB-agar plates supplemented with 100 μg/mL ampicillin
Plasmid miniprep kit (e.g., Qiagen, catalog number: 27104)
Plasmid Plus Midi kit (e.g., Qiagen, catalog number: 12941)
Sequencing primers: pTTfwd 5’ TCCACAGGTGTCCACTCC 3’
pTTrev 5’ TCCTTTATTAGCCAGAGG 3’
Expression in expi293TM cell
500 mL sterile baffled flasks with vented closure (ThermoFisher Scientific, catalog number: 4116-0500)
CountessTM cell counting chamber slides (ThermoFisher Scientific, catalog number: C10228)
Expi293TM cells (ThermoFisher Scientific, catalog number: A14527)
Expi293TM expression medium (ThermoFisher Scientific, catalog number: A1435101)
Trypan blue solution (ThermoFisher Scientific, catalog number: 15250061)
Hanks' Balanced Salt Solution (HBSS) (10×) (ThermoFisher Scientific, catalog number: 14185052)
Gibco OPTI-MEM reduced serum medium (ThermoFisher Scientific, catalog number: 31985062)
Valproic acid (Sigma-Aldrich, catalog number: P4543)
Glucose (45% solution) (see Recipes or from Sigma-Aldrich, catalog number: G8769)
Sodium propionate (Sigma-Aldrich, catalog number: P1880)
PEI MAX 40 K (Polysciences Inc., catalog number: 24,765-1)
Kifunensine (Sigma-Aldrich, catalog number: K1140)
Pre-cast SDS polyacrylamide gels (e.g., NuPAGETM 10%, Bis-Tris ThermoFisher Scientific, catalog number WG1201A)
InstantBlue® Coomassie protein stain (Abcam, catalog number: ab119211)
RBD purification
5 ml HisTrap_FF Ni-NTA columns (Cytiva, catalog number: 1752860)
SD75 16/600 size exclusion column (Cytiva, catalog number: 28989333)
Imidazole (Sigma-Aldrich, catalog number: I2399)
PBS, Phosphate Buffered Saline, 10× Solution (Fisher, catalog number: BP399-20)
Wash buffer (see Recipes)
Elution buffer (see Recipes)
Gel filtration buffers (see Recipes)
VHH production
Vector construction
pADL-23c vector cut with SfiI (Figure 1B) (New England Biolabs, catalog number: R0123S)
Infusion primers:
VHH Fwd primer 5’ GTTATTACTCGCGGCCCAGCCGGCCATGGCCC 3’
VHH Rev primer 5’ GGTGATGGTGTTGGCCTTTATTAATGATGGTGGTGATGGTG 3’
Proof reading polymerase (e.g., PhusionFlashTM high fidelity polymerase ThermoFisher Scientific, catalog number: F548S)
pADL-23c sequencing primers: PhDseqFwd 5’ GCTTCCGGCTCGTATGTTG 3’
PhDseqRev 5’ GTCGTCTTTCCAGACGTTAG 3’
2 mL Cryovials (e.g., ThermoFisher Scientific, catalog number: 5000-0020)
E. coli expression
WK6 cells Escherichia coli (Migula) Castellani and Chalmers (ATCC, catalog number: 47078)
Terrific Broth (TB) Medium (see Recipes)
Isopropyl β-D-1-thiogalactopyranoside, IPTG (Sigma-Aldrich, catalog number: I6758)
Glucose
Magnesium chloride
Ampicillin
VHH purification
TES buffer (see Recipes)
DNase I (Sigma-Aldrich, catalog number: D4263)
Magnesium Chloride (Sigma-Aldrich, catalog number: M8266)
Sucrose (Sigma-Aldrich, catalog number: S0389)

Figure 1. Vector maps (A) pOPINTTGneo (B) pADL-23c
Crystallization of VHH-RBD complexes
Preparation of VHH-RBD complexes
EndoH ( New England Biolabs, catalog number: P0702S)
SD200 10/300 size exclusion column (Cytiva, catalog number: 28990944)
Crystallization screening
Swisssci Triple-Drop crystallization plates (Molecular Dimensions, catalog number: MD11-003-100)
V well Microplate without lid, natural, polypropylene (Greiner, catalog number: 651201 96)
VIEWsealTM plate sealer, transparent, non-piercable Greiner, catalog number: 676070)
Pact premierTM crystallization screen condition: (Molecular Dimension, catalog number: MDSR-29)
JCSG-plusTM crystallization screen condition: (Molecular Dimensions, catalog number: MDSR-37)
SG1TM crystallization condition: (Molecular Dimensions, catalog number: MDSR-88)
Cryopreservation of crystals
Glycerol (Molecular Dimensions, catalog number: MD2-100-65)
PEG 400 (Sigma-Aldrich, catalog number: 06855)
Mounted Round LithoLoops (0.25 mm) (Molecular Dimensions, catalog number: MD7-137)
Mounted Round LithoLoops (0.15 mm) (Molecular Dimensions catalog number: MD7-135)
Standard Foam Dewar (Molecular Dimensions, catalog number: MD7-35)
Uni-Puck 10 Pack ( Molecular Dimensions, catalog number: MD7-613)
Cryotool set (Molecular Dimensions, catalog number: MD7-517)
Dry Shipper (Molecular Dimensions, catalog number: MD7-21
Equipment
Microvolume spectrophotometer (e.g., ThermoFisher Scientific, NanoDropTM One/OneC Microvolume UV-Vis Spectrophotometer: ND-ONE-W)
CO2 orbital shaker (e.g., n-Biotek.com ANICELL incubator, catalog number: NB-206CXL/NB)
Tabletop centrifuge suitable for 50 mL Falcon tubes (Sorvall Legend RT Plus)
Cell Counter (e.g., ThermoFisher Scientific CountessTM 3 Automated Cell Counter)
ÄKTA Xpress automated multi-step purification system
Liquid handler (e.g., Hydra Dispenser by Art Robbins Instruments)
Low volume dispenser for crystallization plates (e.g., SPT labtech mosquito® LV)
Crystal plate imager (e.g., Formulatrix RockImager® system)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Le Bas, A., Mikolajek, H., Huo, J., Norman, C., Dormon, J., Naismith, J. H. and Owens, R. J. (2022). Production and Crystallization of Nanobodies in Complex with the Receptor Binding Domain of the SARS-CoV-2 Spike Protein. Bio-protocol 12(9): e4406. DOI: 10.21769/BioProtoc.4406.
分类
生物物理学 >
生物化学 > 蛋白质 > 表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link








