- EN - English
- CN - 中文
An Improved EMSA-based Method to Prioritize Candidate cis-REs for Further Functional Validation
一种改进的基于EMSA的方法来优先考虑候选的顺式RE,以进一步验证其功能
发布: 2022年04月20日第12卷第8期 DOI: 10.21769/BioProtoc.4397 浏览次数: 2867
评审: Alessandro DidonnaJoyce ChiuAbhilash Padavannil
Abstract
Cells are the complex product of gene expression programs that involve the coordinated transcription of thousands of genes controlled by cis-regulatory elements (cis-REs). Therefore, identification of cis-REs is the key to decipher the mechanisms underlying the regulation of gene expression. Here, we describe a simple and time-effective protocol of fine-mapping cis-REs by using an electrophoresis mobility shift assay (EMSA)-based, in vitro, high-throughput (HTP) technique called regulatory element-sequencing (Reel-seq). Reel-seq can be applied to identify cis-REs at a high resolution and sensitivity over large genome regions, in a systematic and continuous manner. It can be used to prioritize candidate cis-REs as a complement to the existing approaches, such as massive parallel reporter assay (MPRA), chromatin immunoprecipitation DNA-sequencing (ChIP-seq), and the assay for transposase-accessible chromatin sequencing (ATAC-seq).
Graphical abstract:
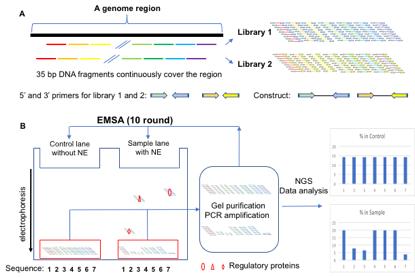
Generation of the Reel-seq Library 1 and 2 (A) and identification of cis-REs by an electrophoresis mobility shift assay (EMSA)-based Reel-seq screen (B). NE: nuclear extract; NGS: next generation sequencing.
Background
Genes make proteins through two steps: transcription and translation. However, specific expression of genes in particular cells is largely regulated by transcription, which is, in general, controlled by millions of promoters and enhancers. Both promoters and enhancers contain short DNA sequences that serve as cis-regulatory elements (cis-Res), which regulate gene transcription by recruiting regulatory proteins (Mikhaylichenko et al., 2018; Schoenfelder and Fraser, 2019). In addition, gene transcription can also be regulated on the epigenetic level, by opening the chromatin structure to alter DNA accessibility, thereby making the underlying cis-REs accessible to regulatory proteins (Crawford et al., 2006; Angeloni and Bogdanovic, 2019; Zhang et al., 2021). Therefore, identification of cis-REs is the key for understanding of biology.
To date, a number of high-throughput (HTP) approaches have been developed to identify cis-REs. Some identify cis-REs directly, by determining the functional impact of each DNA fragment using HTP reporter assays, such as massive parallel reporter assay (MPRA) (Ulirsch et al., 2016; Tewhey et al., 2016), multiplexed editing regulatory assay (MERA) (Rajagopal et al., 2016), clustered regularly interspaced short palindromic repeats interference, flow cytometry and RNA fluorescence in situ hybridization (CRISPRi-FlowFISH) (Fulco et al., 2019), and cis-regulatory element scan by tiling-deletion and sequencing (CREST-seq) (Diao et al., 2017). Some use antibodies specifically against transcriptional factors or epigenetic markers, to identify cis-REs based on protein-DNA interactions, such as chromatin immunoprecipitation DNA-sequencing (ChIP-seq) (Park et al., 2009), and chromatin immunocleavage sequencing (ChIC-seq) (Schmid et al., 2004). Others use the open chromatin regions that are indicative of active regulatory sites, to identify cis-Res, such as DNase-seq (Song and Crawford, 2010), and the assay for transposase-accessible chromatin sequencing (ATAC-seq) (Buenrostro et al., 2015). While all these approaches have their advantages and specific applications in identifying cis-REs, the technical complexity of these methods limits their routine application. In addition, some of these approaches do not have the high resolution to define each single cis-RE, whereas others do not have enough sensitivity and fidelity to reveal cis-REs. Recently, we developed a simple and time-effective technique that not only complements the pre-existing approaches, but can also be used to prioritize candidate cis-REs (Wu et al., 2022). We refer to this technique as regulatory element-sequencing (Reel-seq). Reel-seq is an in vitro HTP screen technique, designed to identify cis-REs based on protein-DNA interaction that is detected by an electrophoresis mobility shift assay (EMSA). For screening, a Reel-seq construct will be engineered by placing a 35-bp DNA fragment between two primers for PCR amplification, as well as for next-generation sequencing (see Graphic abstract A). A Reel-seq library (e.g., Library 1) will be generated by hundreds of thousands of these constructs synthesized by massive parallel oligonucleotide synthesis, so that an entire DNA region will be reconstructed. To cover the break points between all 35-bp fragments, another Reel-seq library (e.g., Library 2) will be generated in the same fashion, except that each 35-bp fragment in this library will bridge the break point of the two contiguous 35-bp fragments in Library 1. For screening, each Reel-seq library will be first mixed with and without nuclear extract (NE, source of regulatory proteins) isolated from relevant types of cells or tissues, and then analyzed on a TBE native gel. After electrophoresis, unshifted bands from both the buffer-treated control and the NE-treated samples will be isolated, and amplified by PCR. Amplified DNA will then be used for another round of gel shifting. In total, ten rounds of EMSA will be performed so that, if a DNA fragment in the library is a cis-RE, it will be shifted in each round of the gel shift assay. After ten screening rounds, a cis-RE can be identified by its decreased percentage in the unshifted DNA library (see Graphic abstract B). Since Reel-seq is an EMSA-based assay, it is highly specific and sensitive in identifying cis-REs. Moreover, since Reel-seq identifies cis-REs at a high resolution, within a defined 35-bp fragment, each identified cis-RE can be used as a “bait” to pull down the regulatory proteins, by using flanking restriction enhanced DNA pulldown-mass spectrometry (FREP-MS) (Li et al., 2018; Wu et al., 2022), another technique recently developed in our lab. Here, we present the detailed protocol of Reel-seq, for the convenient application of this method to identify cis-REs.
Materials and Reagents
150 × 21 mm Dish, NunclonTM Delta (Thermo Fisher, catalog number: 168381)
AccuPrimeTM Taq DNA Polymerase System (Thermo Fisher, catalog number: 12339016. Upon receipt, store at -20°C)
Gel Loading Dye, Blue (6×) [New England Biolabs, catalog number: B7021S. Upon receipt, store at room temperature (RT)]
5% Mini-PROTEAN® TBE Gel, 10 well (Bio-Rad, catalog number: 4565014. Upon receipt, store at 4°C)
PierceTM 10× TBE Buffer (Thermo Fisher, catalog number: 28355. Upon receipt, store at RT)
TE buffer (Thermo Fisher, catalog number: 12090015. Upon receipt, store at RT)
10,000× GelStarTM Nucleic Acid Gel Stain (Lonza, catalog number: 50535. Upon receipt, store at -20°C)
Low molecular weight DNA ladder (New England Biolabs, catalog number: N3233S. Upon receipt, store at -20°C)
NE-PERTM Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher, catalog number: 78833. Upon receipt, store at 4°C)
Roche cOmplete, Mini Protease Inhibitor Cocktail (25 tablets) (Emerald Scientific, catalog number: 11836153001. Upon receipt, store at 4°C)
LightShiftTM Chemiluminescent EMSA Kit (Thermo Fisher, catalog number: 20184. Upon receipt, store kit components at -20°C, except for the Chemiluminescent reagents, which are stored at 4°C as described by the manufacturer)
Herculase II Fusion DNA Polymerases (Agilent, catalog number: 600677. Upon receipt, store at -20°C)
UltraPureTM Agarose (Thermo Fisher, catalog number: 16500100. Upon receipt, store at RT)
Safe-Red (APPLIED BIOLOGICAL MATERIALS INC, catalog number: G108-R. Upon receipt, store at -20°C)
All primers are purchased from Integrated DNA Technologies (IDT) as a standard formula
GibcoTM DPBS, no calcium, no magnesium (Gibco, catalog number: 14190235. Upon receipt, store at RT)
Primary human arterial ECs (Lonza, catalog number: CC-2535)
EGMTM-2 Endothelial Cell Growth Medium-2 BulletKitTM (Lonza, catalog number: CC-3162)
0. 5× TBE buffer (see Recipes)
1× GelStar (see Recipes)
1× Low Molecular Weight DNA Ladder (see Recipes)
10% TE buffer (see Recipes)
1.5% agarose gel (see Recipes)
Equipment
T100 Thermal Cycler (Bio-Rad, catalog number: 1861096)
PowerPacTM Basic Power Supply (Bio-Rad, catalog number: 1645050)
Mini-PROTEAN® Tetra Vertical Electrophoresis Cell (Bio-Rad, catalog number: 1658003FC)
Eppendorf ThermoMixerTM C (Fisher Scientific, catalog number: 13527550)
Thermo Scientific Savant DNA 120 SpeedVac Concentrator (Thermo Scientific, catalog number: BZ10133848)
BLooKTM LED Transilluminator (GeneDireX, catalog number: BK001)
Implen C40 NanoPhotometer UV/Vis Spectrophotometer Mobile System, 110/220 V (Cole-Parmer, catalog number: UX-83070-62)
Cell scraper (VWR, catalog number: 15621-005)
VWR Ergonomic High-Performance Pipettor set (VWR, catalog number: 89079-970)
Thermo Forma Steri Cycle 370 CO2 Incubator (Marshall Scientific, catalog number: TH-370)
Allegra® X-5 Clinical Benchtop IVD Centrifuge, Beckman Coulter® (VWR, catalog number: 89429-566)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Wu, T. and Li, G. (2022). An Improved EMSA-based Method to Prioritize Candidate cis-REs for Further Functional Validation. Bio-protocol 12(8): e4397. DOI: 10.21769/BioProtoc.4397.
分类
分子生物学 > DNA > 基因表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link