- EN - English
- CN - 中文
RNA Interference Method for Gene Function Analysis in the Japanese Rhinoceros Beetle Trypoxylus dichotomus
日本犀牛甲虫Trypoxylus dichotomus基因功能分析的RNA干扰方法
(*contributed equally to this work) 发布: 2022年04月20日第12卷第8期 DOI: 10.21769/BioProtoc.4396 浏览次数: 2550
评审: Teiya KijimotoLeonardo Gaston GuilgurAnonymous reviewer(s)
Abstract
In the Japanese rhinoceros beetle Trypoxylus dichotomus, various candidate genes required for a specific phenotype of interest are listed by next-generation sequencing analysis. Their functions were investigated using RNA interference (RNAi) method, the only gene function analysis tool for T. dichotomus developed so far. The summarized procedure for the RNAi method used for T. dichotomus is to synthesize double-stranded RNA (dsRNA), and inject it in larvae or pupae of T. dichotomus. Although some dedicated materials or equipment are generally required to inject dsRNA in insects, the advantage of the protocol described here is that it is possible to inject dsRNA in T. dichotomus with one syringe.
Keywords: RNA interference (RNA干扰)Background
The Japanese rhinoceros beetle Trypoxylus dichotomus is useful from different perspectives, such as in developmental biology, morphology, ethology, ecology, biomimetics, aerodynamics, and drug discovery. For instance, Siva-Jothy (1987) revealed the feeding time of T. dichotomus from ecological research, Miyanoshita et al. (1996) isolated a new antibacterial peptide from larvae of T. dichotomus, Hongo (2007) clarified the relationship between horn length and body size in T. dichotomus, Emlen et al. (2012) showed that signaling through the insulin receptor is involved in T. dichotomus horn development, Chen et al. (2017) clarified that the elytron plate of T. dichotomus has a mechanism of high energy absorption, and Ohde et al. (2018) identified several horn formation genes using RNA-sequencing analysis and RNAi techniques.
Recently, the genome of T. dichotomus was clarified (Ogata, 2021; Morita et al., 2022), and various candidate genes required for a specific phenotype of interest were listed by RNA-sequencing analysis (Ohde et al., 2018; Zinna et al., 2018). Furthermore, the functions of the candidate genes were investigated using RNA interference (RNAi) method, the only gene function analysis tool for T. dichotomus developed so far (Emlen et al., 2012; Ito et al., 2013; Gotoh et al., 2015; Adachi et al., 2018, 2020; Ohde et al., 2018; Morita et al., 2019).
RNAi method for small insects generally requires specialized setups for microinjection, such as those for preparing glass capillaries, loading double-stranded RNA (dsRNA) into microcapillaries, injecting dsRNA into insects using high pressure, and manipulating the needle for injection under a stereomicroscope (e.g., Kim et al., 2004; Niimi et al., 2005; Masumoto et al., 2009; Posnien et al., 2009). The RNAi method in T. dichotomus does not require most of the above setups, it only requires a 1-mL syringe for dsRNA injection, since the beetle’s larval body size is as large as a mouse. This study provides a detailed protocol for the RNAi method in the third instar larvae of T. dichotomus (Figure 1). This method can also be applied to the first and second instar larvae and pupae of T. dichotomus.
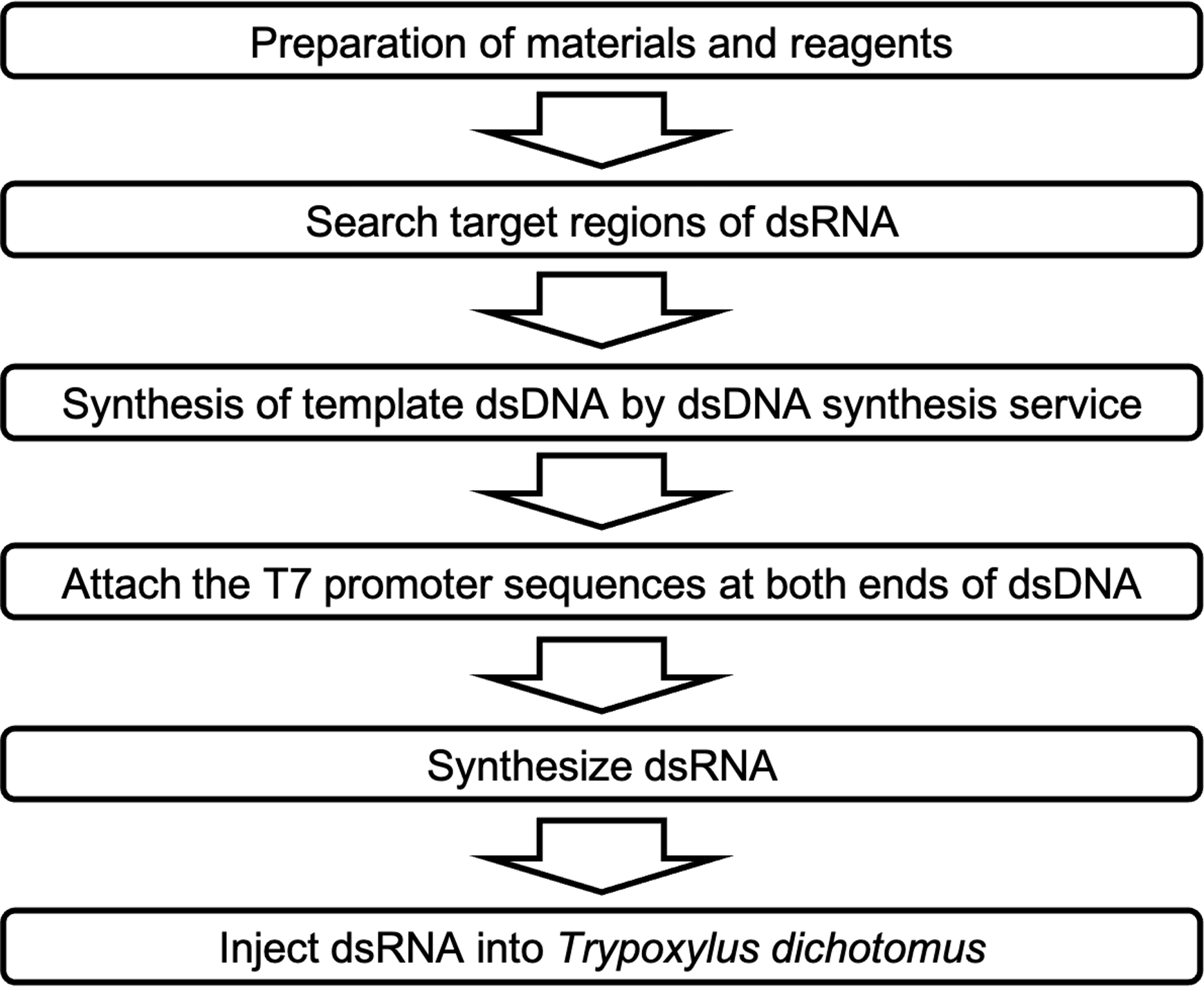
Figure 1. Overview of RNA interference method procedure in Trypoxylus dichotomus.
Materials and Reagents
Filter Pipette tips (RNase/DNase-free) (Labcon, catalog numbers: 1057-965-018-9 [P1000], 1059-965-008-9 [P200], 1055-965-018-9 [P40]; and QSP, catalog number: TF102-10-Q [P2])
Disposable needles No.30 (30-gauge) (Dentronics, No.30)
0.2 mL Flat PCR Tube 8-Cap Strips (INA OPTIKA, catalog number: 3247-00)
1.5 mL microtubes, flat-bottom, DNase/RNase-free (Watson, catalog number: 131-415C)
Plastic bottles (Hobby Club, blow container 750)
Lavender Nitrile Powder-free Exam Gloves (Kimberly-Clark, catalog number: 52818)
Styrofoam container
Aluminum foil
Third instar larvae of Trypoxylus dichotomus (Loiinne, third instar larva)
UltraPureTM DNase/RNase-Free Distilled Water (UPW) (Invitrogen, catalog number: 10977023)
Loading Dye (New England Biolabs, catalog number: B7024S)
100 bp DNA Ladder (New England Biolabs, catalog number: N3231L)
TaKaRa Ex Taq (TaKaRa, catalog number: RR001B)
AmpliScribeTM T7-FlashTM Transcription Kit (Epicentre Technologies, catalog number: ASF3257)
Rnase AWAY (Thermo Fisher Scientific, catalog number: 7002)
Monarch PCR & DNA Cleanup Kit (5 μg) (BioLabs, catalog number: T1030)
Terumo Syringe 1 mL for Tuberculin Slip Tip White (Terumo, catalog number: SS-01P)
Ethanol (FUJIFILM Wako Chemicals, catalog number: 057-00456)
Phenol/Chloroform/Isoamyl alcohol (25:24:1) (PCI) (NIPPON GENE, catalog number: 311-90151)
Chloroform/Isoamyl alcohol (24:1) (CIA) (Sigma-Aldrich, catalog number: C0549)
3 M Sodium Acetate (pH 5.2) (NIPPON GENE, catalog number: 316-90081)
Agarose S (NIPPON GENE, catalog number: 318-01195)
10× TAE Buffer (NIPPON GENE, catalog number: 318-90301)
Ethidium Bromide Powder (SIGMA, catalog number: E7637-1G)
Humus (Dorcus Owner’s shop, Osaka, Japan)
PCR Reaction mixture (see Recipes)
Solution for 1% agarose gel (see Recipes)
Equipment
Thermal cycler (Bio-Rad Laboratories, model: T100)
Pipetman (GILSON, catalog numbers: F123602 [P1000], F123601 [P200], F123600 [P20], and F144801 [P2])
Spectrophotometer (DeNovix, model: DS-11)
Electrophoresis chamber (TaKaRa, model: AD115)
Microwave oven (Panasonic, model: NE-MS261)
Gel imaging device (ATTO, model: WSE-6100)
Centrifuge (TOMY, model: MX-307)
Heat block (Major Science, model: MD-MINI)
Vortex mixer (LMS, model: VTX-3000L)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Sakura, K., Morita, S. and Niimi, T. (2022). RNA Interference Method for Gene Function Analysis in the Japanese Rhinoceros Beetle Trypoxylus dichotomus. Bio-protocol 12(8): e4396. DOI: 10.21769/BioProtoc.4396.
分类
发育生物学 > 形态建成 > 变态发育
分子生物学 > RNA > RNA 干扰
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link