- EN - English
- CN - 中文
An Optimized Tat/Rev Induced Limiting Dilution Assay for the Characterization of HIV-1 Latent Reservoirs
一种优化的 Tat/Rev 诱导的限制稀释测定表征 HIV-1 潜伏储库
(*contributed equally to this work) 发布: 2022年04月20日第12卷第8期 DOI: 10.21769/BioProtoc.4391 浏览次数: 3014
评审: Alessandro DidonnaNarayan SubramanianSalim Gasmi
Abstract
The administration of antiretroviral therapy (ART) leads to a rapid reduction in plasma viral load in HIV-1 seropositive subjects. However, when ART is suspended, the virus rebounds due to the presence of a latent viral reservoir. Several techniques have been developed to characterize this latent viral reservoir. Of the various assay formats available presently, the Tat/Rev induced limiting dilution assay (TILDA) offers the most robust and technically simple assay strategy. The TILDA formats reported thus far are limited by being selective to one or a few HIV-1 genetic subtypes, thus, restricting them from a broader level application. The novel TILDA, labelled as U-TILDA ('U' for universal), can detect all the major genetic subtypes of HIV-1 unbiasedly, and with comparable sensitivity of detection. U-TILDA is well suited to characterize the latent reservoirs of HIV-1 and aid in the formulation of cure strategies.
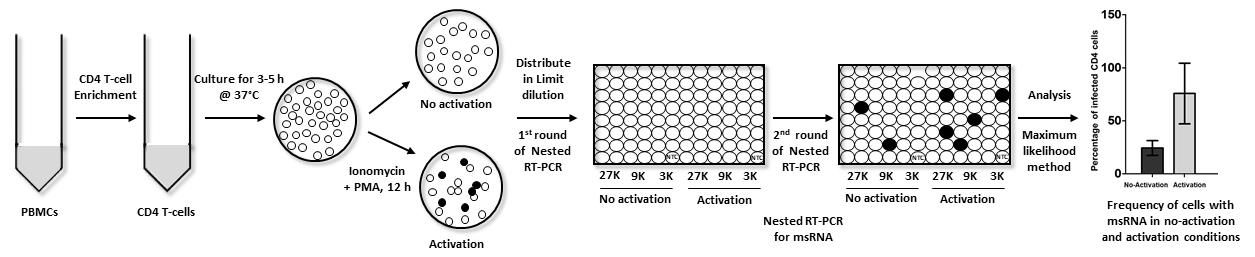
Graphical abstract:
Background
Latent reservoir (LR) poses a significant obstacle to HIV-1 disease management. Despite successful antiretroviral therapy (ART), low-level viral replication persists, attributed to the existence of a transcriptionally silent virus (Chun et al., 1995, 1997; Finzi et al., 1997; Siliciano et al., 2003). Precise quantitation of the size of this latent reservoir before and after the medical intervention is critical for 'cure' or therapeutic research.
A variety of assay formats have been developed to characterize HIV-1 latent reservoirs (Falcinelli et al., 2019). These assays differ from one another in terms of the viral product they measure, complexity, and sensitivity, among other attributes. DNA PCR-based assays offer the technically simplest experimental format. However, such assays quantify total proviral DNA, regardless of replication competence of the integrated viruses, thus, overestimating the overall viral reservoir size (Massanella and Richman, 2016). The highly popular quantitative viral outgrowth assay (QVOA) measures the number of infectious viral units per million CD4 T-cells (Siliciano and Siliciano, 2005). The QVOA, considered to be the gold standard technique, suffers from several limitations, including the complex experimental design, and the need for a large quantity of blood (120–180 mL). Additionally, since only a fraction of replication-competent proviruses is activated under any experimental condition, QVOA tends to underestimate the size of the viral reservoir.
Among a horde of assays available to characterize HIV-1 latent reservoirs, a strategy that aims to detect the frequency of cells producing viral transcripts offers the most powerful platform. The Tat/rev induced limiting dilution assay (TILDA) can measure cells producing multiply-spliced viral transcripts following cell activation (Procopio et al., 2015). The TILDA format offers a realistic approximation of the size of the replication-competent latent reservoir, as it detects HIV-1 transcripts, not proviral DNA. Importantly, TILDA exploits the power of PCR amplification, thus offering a highly sensitive experimental format. Additionally, unlike QVOA, TILDA requires a significantly smaller volume of a clinical sample (15–20 mL). Despite these considerable technical merits, TILDA is highly susceptible to the genetic variation of several HIV-1 viral subtypes. Therefore, the several TILDA platforms reported thus far are limited by the ability to target a specific HIV-1 genetic family, or even only a proportion of viral strains of a subtype (Bertoldi et al., 2020; Leyre et al., 2020). We recently reported a novel TILDA assay format labelled as U-TILDA ('U' for universal), that can target all the major genetic subtypes of HIV-1 (A, B, C, D, and AE subtypes) with high sensitivity and comparable diagnostic efficiency (Mehta et al., 2021). In contrast to subtype-specific TILDA formats, in U-TILDA, the primer pairs and the probe target the highly conserved exons of HIV-1, thus significantly broadening the breadth of detection of the assay.
Here, we describe the detailed protocol of U-TILDA using stored PBMC of an HIV-1 seropositive subject. The CD4 T-cells, negatively enriched using paramagnetic beads, and activated with PMA and ionomycin for 12 h, or left without activation for control, were serially diluted. One step qRT-PCR was performed in sixteen replica wells for each cell dilution. The viral transcripts were converted into cDNA using HIV-1-specific RT primers, without RNA purification. This was followed by the first-round of PCR amplification in the same reaction vial. Diluted PCR reaction content was then used as the template for the second-round of the nested-PCR, in a real-time PCR format. Finally, the Ct values of the replicate wells were converted to the frequency of transcription-competent viral events using a free web tool.
Materials and Reagents
Consumables and reagents
Conical centrifuge tubes, 50 mL (Thermo Fisher Scientific, catalog number: 14-432-22)
Conical centrifuge tubes, 15 mL (Thermo Fisher Scientific, catalog number: 14-959-53A)
Round-bottom polystyrene tubes, 5 mL (Thermo Fisher Scientific, catalog number: 14-959-1A)
Microcentrifuge tubes; 0.5, 1.5, and 2 mL (Tarsons, catalog numbers: 500000, 500010, and 500020 respectively)
PCR plate (Bio-Rad Laboratories, catalog number: HSP9601)
RPMI 1640 medium (HiMedia Laboratories, catalog number: AL162S)
Fetal bovine serum (FBS, Heat-inactivated) (Thermo Fisher Scientific, catalog number: 10082147)
Penicillin G (Sigma-Aldrich, catalog number: P3032)
L-glutamine (Sigma-Aldrich, catalog number: G8540)
Streptomycin (Sigma-Aldrich, catalog number: S9137)
EasySep Human CD4+ T cell isolation kit (Stemcell Technologies Inc., catalog number:19052)
Phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich, catalog number: P1585)
Ionomycin (Sigma-Aldrich, catalog number: I0634)
SuperScript III Platinum One-Step qRT-PCR Kit (Invitrogen, catalog number: 11732020)
MyTaqTM DNA polymerase (Bioline, catalog number: BIO-21105)
PCR sealant (Bio-Rad Laboratories, catalog number: MSB1001)
FITC Mouse Anti-Human CD4 antibody (BD Biosciences, catalog number: 566802)
FITC Mouse IgG1, ĸ Isotype Control (BD Biosciences, catalog number: 349041)
APC Mouse Anti-Human CD3 antibody (BD Biosciences, catalog number: 555342)
APC Mouse IgG2a, ĸ Isotype Control (BD Biosciences, catalog number: 550882)
Live/Dead Fixable Far Red Dead Cell Stain Kit, for 633 or 635 nm excitation (Thermo Fisher Scientific, catalog number: L10120)
Peripheral blood mononuclear cells (PBMC)
Revival media (see Recipes)
Enrichment media (see Recipes)
Staining buffer (see Recipes)
RPMI medium supplemented with 10% FBS (see Recipes)
Phosphate-buffered saline (PBS) solution (see Recipes)
Tris-EDTA (TE) buffer (see Recipes)
40 µg/mL PMA (see Recipes)
4 µg/mL Ionomycin (see Recipes)
Primers and probe (Table 1)
Table 1. Details of the primers and probe used in PCR and cDNA synthesis
Oligonucleotides
Identity
Coordinates (HXB2)
Sequence (5’-3')
Primers
N2830 (OFP)
514-540
CTGCTTAAGCCTCAATAAAGCTTGCCT
N2831 (ORP)
8510-8539
CTCAATCGGTGGTAGCTGAAGAGGCACAGG
N2832 (IFP)
577-603
GACTCTGGTAACTAGAGATCCCTCAGA
N2833 (IRP)
8419-8448
TGTCTTGCTCTCCACCTTCTTCTTCGATTC
Probe
N2842
683-709
(FAM)-CTCTCGACGCAGGACTCGGCTTGCTGA-(BHQ-1)
RT primers
N4101
9031-9046
TTTTTTTTTTTTTTCAGAGCACTC
N4102
9032-9046
TTTTTTTTTTTTTCAGAGCACTCAAG
N4103
9033-9046
TTTTTTTTTTTTTTCAGAGCACTCAAGG
Notes:
OFP and ORP: Outer Forward and Reverse Primers, IFP and IRP: Inner Forward and Reverse Primers, RT primers: Reverse Transcription primers.
The reverse primer sequences are presented in the reverse complement.
The primers were synthesized by Sigma-Aldrich.
Equipment
Liquid Nitrogen container (IOCL Cryocan)
4°C refrigerator (Samsung)
–80°C freezer (Thermo Scientific)
CO2 incubator (NuAire)
Water bath (BS Enterprises)
CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories)
Laminar flow hood (NV-Equipes)
Inverted microscope (Leica DM IRB)
Flow cytometer (BD FACSAriaTM III)
Hemocytometer (TECH- LABCARE)
Software
FCS 6 ExpressTM Flow Cytometry (De NovoTM Software)
GraphPad Prism 5 (GraphPad Software, Inc.)
Extreme Limiting Dilution Analysis (ELDA) (Walter + Eliza Hall Bioinformatics, Institute of Medical Research). A free web tool available at https://bioinf.wehi.edu.au/software/elda/
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Mishra, S., Gohil, Y., Mehta, K., D'silva, A., Amanullah, A., Selvam, D., Pargain, N., Nala, N., Sanjeeva, G. N. and Ranga, U. (2022). An Optimized Tat/Rev Induced Limiting Dilution Assay for the Characterization of HIV-1 Latent Reservoirs. Bio-protocol 12(8): e4391. DOI: 10.21769/BioProtoc.4391.
分类
免疫学 > 免疫细胞分化 > T 细胞
分子生物学 > DNA > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link