- EN - English
- CN - 中文
Tracking the Reversed Oxidative Tricarboxylic Acid Cycle in Bacteria
追踪细菌中的反向氧化三羧酸循环
(*contributed equally to this work) 发布: 2022年03月20日第12卷第6期 DOI: 10.21769/BioProtoc.4364 浏览次数: 4242
评审: Wenrong HeYuan WangYe Xu
Abstract
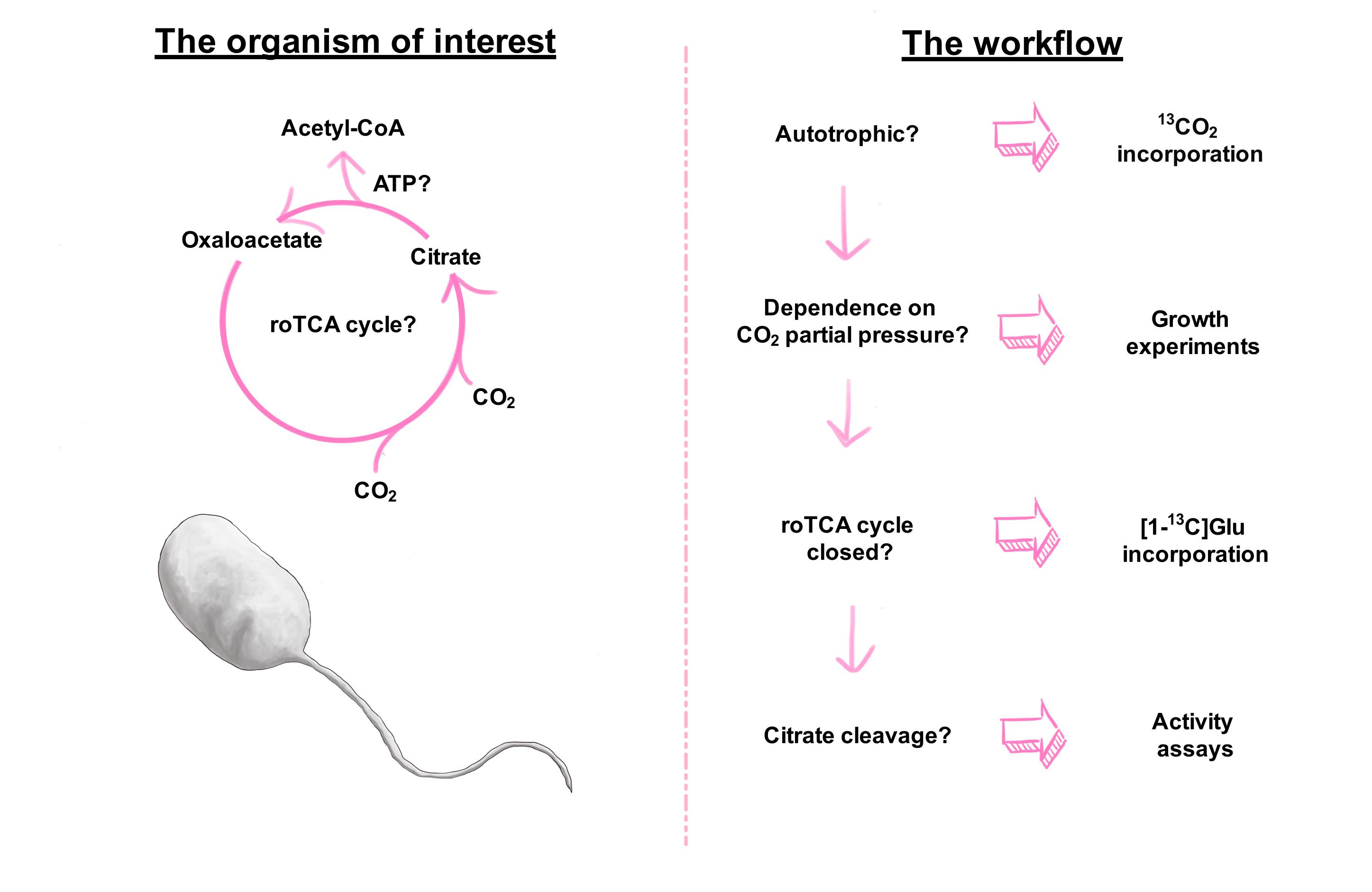
Different pathways for autotrophic CO2 fixation can be recognized by the presence of genes for their specific key enzymes. On this basis, (meta)genomic, (meta)transcriptomic, or (meta)proteomic analysis enables the identification of the role of an organism or a distinct pathway in primary production. However, the recently discovered variant of the reductive tricarboxylic acid (rTCA) cycle, the reverse oxidative tricarboxylic acid (roTCA) cycle, lacks unique enzymes, a feature that makes it cryptic for bioinformatics analysis. This pathway is a reversal of the widespread tricarboxylic acid (TCA) cycle. The functioning of the roTCA cycle requires unusually high activity of citrate synthase, the enzyme responsible for citrate cleavage, as well as elevated CO2 partial pressures. Here, we present a detailed description of the protocol we used for the identification of the roTCA cycle in members of Desulfurellaceae. First, we describe the anaerobic cultivation of Desulfurellaceae at different CO2 concentrations with a method that can be adapted to the cultivation of other anaerobes. Then, we explain how to measure activities of enzymes responsible for citrate cleavage, malate dehydrogenase reaction, and the crucial carboxylation step of the cycle catalyzed by pyruvate synthase in cell extracts. In conclusion, we describe stable isotope experiments that allow tracking of the roTCA cycle in vivo, through the position-specific incorporation of carbon-13 into amino acids. The label is provided to the organism as 13CO2 or [1-13C]glutamate. The same key methodology can be used for the reliable evaluation of the functioning of the roTCA cycle in any organism under study. This pathway is likely to participate, completely unseen, in the metabolism of various microorganisms.
Graphic abstract:
Background
The study of metabolic pathways responsible for the fixation of inorganic carbon is important for the understanding of the evolution of early cellular life (Fuchs, 2011; Weiss et al., 2016) and the investigation of current global issues, such as climate change. Being responsible for the net fixation of 7 × 1016 g carbon annually (Field et al., 1998), this biological process is critical for the sustainment of life on Earth. Until today, seven different autotrophic CO2 fixation pathways have been described (Berg, 2011; Fuchs, 2011; Sánchez-Andrea et al., 2020). In most cases, the detection of genes that encode enzymes catalyzing key steps in a specific pathway in (meta)genomes and (meta)transcriptomes allows us to estimate the contribution of the corresponding organisms or communities to primary production. However, the recent discovery of the reversed oxidative tricarboxylic acid (roTCA) cycle in the obligate anaerobic sulfur reducers, Desulfurella acetivorans (Mall et al., 2018) and Thermosulfidibacter takaii (Nunoura et al., 2018), challenges this view. The roTCA cycle is the reversal of the widespread (Krebs) tricarboxylic acid (TCA) cycle, and its key features are high energetic efficiency and the lack of any unique enzyme. Being a variant of the reductive TCA (rTCA) cycle, the roTCA cycle utilizes citrate synthase, instead of ATP-citrate lyase, to catalyze the citrate cleavage reaction (citrate + CoA → acetyl-CoA + oxaloacetate + H2O). This step is thermodynamically and kinetically unfavorable—it requires an elevated amount of catalyst (i.e., citrate synthase represents 7% of the total protein content in D. acetivorans [Steffens et al., 2021]), and also an unusually high substrate to product ratio (i.e., CoA/acetyl CoA is 93 in D. acetivorans, using the roTCA cycle [Mall et al., 2018], and only 2.3 in Escherichia coli, using the classical TCA cycle [Bennett et al., 2009]).
Considering the almost universal distribution of citrate synthase in bacteria, this pathway may potentially function unnoticed in many autotrophs, making bioinformatical predictions challenging and raising the question on how to identify it (Mall et al., 2018). Our recent study (Steffens et al., 2021), from which this protocol originates, helped to resolve this issue, but also revealed physiological constraints governing the functioning of the roTCA cycle. Therein, for several representatives of the family Desulfurellaceae (i.e., Hippea maritima, D. acetivorans, D. multipotens, D. propionica), we demonstrated that the usage of the roTCA cycle requires high partial pressure of CO2. The growth of these bacteria showed a clear dependence on CO2 concentration, with cells barely growing at partial pressure of CO2 below 10 kPa. In contrast, Desulfobacter hydrogenophilus, which uses the ATP-citrate lyase-dependent variant of the cycle (Schauder et al., 1987), grew equally well at all tested CO2 concentrations (from 2 to 80 kPa) (Steffens et al., 2021). Our data revealed that the pyruvate synthase reaction (acetyl-CoA + CO2 + reduced ferredoxin → pyruvate + CoA + oxidized ferredoxin) was responsible for this CO2 dependence, as this thermodynamically challenging reaction requires low CoA/acetyl-CoA ratio. As already mentioned, the reversal of the citrate synthase reaction (and thus the functioning of the roTCA cycle) is only possible at high CoA and low acetyl-CoA concentrations, making the following acetyl-CoA carboxylation with pyruvate synthase difficult. This unfavorable condition is compensated by the increase in the concentration of CO2, another substrate of the pyruvate synthase reaction, making the assimilation of acetyl-CoA possible, as the product of the roTCA cycle.
With these features in mind, we can now streamline different techniques in a protocol that will permit the verification of the functioning of the roTCA in promising candidate organisms. Anaerobic cultivation experiments prove the growth dependence on different CO2 concentrations. Extremely high activities of the citrate synthase and malate dehydrogenase reactions in cell extracts are not necessary for other metabolic pathways, thus proving the requirement for an increased amount of catalysts to perform the ATP-independent citrate cleavage. The presence in cell extracts of the critical carboxylases of the pathway (pyruvate and 2-oxoglutarate synthases) further confirms the functioning of the roTCA cycle. Finally, isotopologue profiling of amino acids show position-specific incorporations of carbon-13 and prove the operation of the roTCA cycle in growing bacteria. Cultures grown with 13CO2 show an enrichment of fully labelled amino acids, especially of glutamate, thus proving the existence of a functional CO2 fixation pathway. Cultures grown in the presence of traces of [1-13C]glutamate show the specific formation and accumulation of [4-13C]aspartate, evidencing that the roTCA cycle is closed. However, some bacteria are not capable to transport glutamate into the cells; in this case, the glutamate labelling experiment will not be applicable and alternatives might be considered, such as 13C-labelled pyruvate, glutamine, or citrate.
The identification of promising candidate organisms can be carried out with the help of bioinformatics tools; analysis of (meta)genomes could help to exclude assuredly autotrophic organisms a priori, due to the presence of key enzymatic genes of known CO2 fixation pathways. Nevertheless, the presence of other autotrophic enzymes does not make the functioning of the roTCA cycle impossible. For example, D. multipotens uses the roTCA cycle while possessing an apparently functional gene for ATP-citrate lyase (Mall et al., 2018; Steffens et al., 2021). The usage of software Interactive Codon Analysis (INCA) (Supek et al., 2004, 2005) could help to screen organisms in advance. Indeed, codon usage patterns could be used for the prediction of highly expressed genes, and high expression of the citrate synthase gene is crucial for the functioning of this cycle. However, the results of the codon usage analysis are not decisive and may lead to false-positives or false-negatives, requiring biochemical assays with cell extracts, or perhaps, quantitative proteomics/transcriptomics data for further proof. Accordingly, every case has to be necessarily validated experimentally.
Being an energetically efficient autotrophic pathway, the roTCA cycle may be widespread in natural environments at elevated partial pressures of CO2 (e.g., deep-sea hydrothermal vents) (Steffens et al., 2021). Its elusiveness compromises the reliability of bioinformatics analysis, and the possibility to make solid predictions about the capability of microorganisms for autotrophic growth or primary production. The protocol presented here allows an unambiguous identification of the roTCA cycle in organisms under study.
Materials and Reagents
Cultivation experiments of anaerobic organisms
Serum bottle N20, 120 mL (MediPac, catalog number: 10113100) or equivalent
Infusion bottles, 1 L (VWR, catalog number: 215-9243) or equivalent
Butyl rubber stopper N20 (Glasgerätebau Ochs, catalog number: 102049) or equivalent
Rubber stoppers for 1 L infusion bottles (ERIKS)
Crimp aluminum caps (WICOM, catalog number: WIC 44580) or equivalent
Crimper (VWR, catalog number: 548-0073)
Decapper (VWR, catalog number: 548-0074)
Screw top, with hole Ø 19 mm (VWR, catalog number: MUEL14.076.04)
Duran® flasks, 1 L (VWR, catalog number: SCOT818015403)
Rubber stoppers for Duran® flasks (Glasgerätebau Ochs, catalog number: 444704)
Lecture-bottle control valve (Sigma, catalog number: Z146951)
Sterile syringes, male Luer-Lock (Fisher Scientific, catalog number: 14955461) or equivalent
Luer-to-tubing connector, female Luer-Lock (Sigma, catalog number: Z261580)
Sterile ERSTA syringes for single use, 1 mL (Th. Geyer, catalog number: 6075936) or equivalent
Disposable needles (Carl Roth, catalog number: X132.1) or equivalent
Sterile syringe filters, 0.2 µm pore size (VWR, catalog number: 514-0061)
Na2S·9H2O (Sigma, catalog number: 1313-84-4); store at 4°C
Sulfur powder (Carl Roth, catalog number: 9304.1); store at room temperature (RT)
Resazurin sodium salt (Sigma, catalog number: R7017); store at RT
Hydrogen gas (Air Liquide Deutschland, catalog number: P0231L50R2A001)
Carbon dioxide gas (Air Liquide Deutschland, catalog number: P0760L50R0A001)
Carbon-13 dioxide gas, 99% 13C (Sigma, catalog number: 364592)
L-Glutamic acid-1-13C, 99% 13C (Sigma, catalog number: 604968)
H. maritima medium (modified from DSMZ 854) (see Recipe 1)
D. acetivorans, D. multipotens, and D. propionica medium (modified from DSMZ 480) (see Recipe 1)
SL-10 trace element solution (see Recipe 2)
Wolfe vitamin solution (see Recipe 3)
Trace element solution (DSMZ 141) (see Recipe 4)
Harvesting of biomass and preparation of cell extracts
Folded filters, Ø 240 mm (Whatman Schleicher & Schuell, catalog number: 10311651) or equivalent
Funnels with short stem (VWR, catalog number: 221-0180) or equivalent
Centrifuge bottles with sealing closure, 1 L (Thermo Fisher Scientific, catalog number: 05-564-26) or equivalent
Centrifuge tubes, 50 mL (VWR, catalog number: 21008-940) or equivalent
Microcentrifuge tubes, 1.5 and 2 mL (VWR, catalog number: 72.690.001, 72.689) or equivalent
Serological pipettes (Thermo Fischer Scientific, catalog number: 170350) or equivalent
Liquid nitrogen
0.9% [w/v] NaCl solution (saline)
Aluminum cooling block (Sigma, catalog number: Z743486)
Cell lysis buffer (see Recipe 5)
Enzyme assays
Reaction mixtures are listed. While working, we recommend keeping all chemicals and helping enzymes on ice. All listed components were solved or diluted with dH2O, unless mentioned otherwise. When special attention is required due to their stability, recommended storage conditions or special notes are reported (e.g., freshly solved before the assays). The pH of organic acids and ATP solutions should be adjusted to neutral; the pH of Tris-HCl buffers can be adjusted at RT, but please note that when the final test temperature differs from RT, the pH also changes according to the following formula: ΔpKa/°C≈-0.028 (e.g., the pH of Tris solutions decreases 0.028 pH units for each 1°C increase in temperature). For long-term storage, organic compounds and CoA esters solutions should be kept at -20°C or -80°C, if not mentioned otherwise; all buffers and salt solutions could be kept at RT. Acetyl-CoA was chemically synthesized from the corresponding anhydrides and CoA, according to a previously described method (Simon et al., 1953).
In our case, the concentrations of stock solutions were as follows:
Citrate synthase (forward reaction, spectrophotometric continuous assay)
1 M Tris-HCl (pH 7.5) (Carl Roth, catalog number: 9090.4)
50 mM oxaloacetate, store at -20°C or -80°C for ≤2 months (Sigma, catalog number: 04126)
10 mM acetyl-CoA
10 mM 5,5-dithiobis-2-nitrobenzoic acid (DTNB), freshly solved in 100 mM Tris-HCl (pH 7.5) (Sigma, catalog number: D8130)
Citrate synthase (backward reaction, spectrophotometric continuous assay)
1 M Tris-HCl (pH 7.8) (Carl Roth, catalog number: 9090.4)
50 mM MgCl2
50 mM dithioerythritol (DTE), freshly solved (Sigma, catalog number: D8255)
5 mM CoA (Neofroxx, catalog number: 2100GR005)
200 mM citrate (Sigma, catalog number: C8532)
5 mM NADH, freshly solved (Acros organics, catalog number: 10711911)
200 U/mL porcine malate dehydrogenase, store at 4°C, freshly diluted (Sigma, catalog number: M1567)
Citrate synthase (backward reaction, UHPLC discontinuous assay)
1 M Tris-HCl (pH 7.8) (Carl Roth, catalog number: 9090.4)
50 mM MgCl2
50 mM DTE, freshly solved (Sigma, catalog number: D8255)
5 mM CoA (Neofroxx, catalog number: 2100GR005)
200 mM citrate (Sigma, catalog number: C8532)
5 mM NADH, freshly solved (Acros organics, catalog number: 10711911)
200 U/mL porcine malate dehydrogenase, store at 4°C, freshly diluted (Sigma, catalog number: M1567)
Vial insert, 250 µL, pulled point glass (Agilent, catalog number: 5183-2085)
Screw top vials, 2 mL, and screw caps (Agilent) or equivalent
Microplate, sterile, 96 well (Greiner Bio-One, catalog number: 655161) or equivalent
Membrane filter, 0.22 µm pore size (Sigma, catalog number: GSWP04700)
Acetonitrile anhydrous, 99.8% (Sigma, catalog number: 271004)
Stop solution (see Recipe 6)
UHPLC-grade double distilled water (see Recipe 7)
Potassium phosphate buffer (see Recipe 7)
ATP-citrate lyase
1 M Tris-HCl (pH 7.5) (Carl Roth, catalog number: 9090.4)
100 mM MgCl2
10 mM ATP, freshly solved (Sigma, catalog number: A26209)
50 mM DTE, freshly solved (Sigma, catalog number: D8255)
50 mM CoA (Neofroxx, catalog number: 2100GR005)
100 mM citrate (Sigma, catalog number: C8532)
5 mM NADH, freshly solved (Acros Organics, catalog number: 10711911)
200 UF/mL porcine malate dehydrogenase, store at 4°C, freshly diluted (Sigma, catalog number: M1567)
Citrate lyase
1 M Tris-HCl (pH 7.5) (Carl Roth, catalog number: 9090.4)
100 mM MgCl2
50 mM DTE, freshly solved (Sigma, catalog number: D8255)
100 mM citrate (Sigma, catalog number: C8532)
5 mM NADH, freshly solved (Acros Organics, catalog number: 10711911)
200 U/mL porcine malate dehydrogenase, store at 4°C, freshly diluted (Sigma, catalog number: M1567)
Malate dehydrogenase
1 M Tris-HCl (pH 8) (Carl Roth, catalog number: 9090.4)
50 mM MgCl2
50 mM DTE, freshly solved (Sigma, catalog number: D8255)
25 mM oxaloacetate, store at -20°C or -80°C for ≤2 months (Sigma, catalog number: 04126)
5 mM NADH, freshly solved (Acros organics, catalog number: 10711911)
Pyruvate synthase
1 M Tris-HCl (pH 7.5) (Carl Roth, catalog number: 9090.4)
100 mM MgCl2
50 mM DTE, freshly solved (Sigma, catalog number: D8255)
50 mM CoA (Neofroxx, catalog number: 2100GR005)
100 mM pyruvate (Sigma, catalog number: P2256)
10 mM methyl viologen (Sigma, catalog number: 856177)
2-Oxoglutarate synthase
1 M Tris-HCl (pH 7.5) (Carl Roth, catalog number: 9090.4)
100 mM MgCl2
50 mM DTE, freshly solved (Sigma, catalog number: D8255)
50 mM CoA (Neofroxx, catalog number: 2100GR005)
100 mM 2-oxoglutarate (Sigma, catalog number: 75890)
20 mM benzyl viologen (Sigma, catalog number: D8130)
Isotopologue profiling (based on GC-MS analysis)
Glass vials, 1.5 mL (VWR, catalog number: 548-0018)
Screw caps for GC/MS vials (VWR, catalog number: 548-0814)
Glass inlets, 200 µL (VWR, catalog number: 548-0780)
Microcentrifuge tubes, 2 mL (VWR, catalog number: 525-0228)
Glass Pasteur pipettes, short form (Glasswarenfabrik Karl Hecht GmbH & Co. KG, catalog number: 40567001)
Glass wool, superfine (Glasswarenfabrik Karl Hecht GmbH & Co. KG, catalog number: 41408003)
Hydrochloric acid, 37% (Sigma, catalog number: 320331)
Acetic acid, ≥99% (Sigma, catalog number: A6283)
Dowex 50WX8 200-400 (H±) (Alfa Aesar, catalog number: L13922.18)
Methanol, 99.8% (Sigma, catalog number: 322415)
Ammonia solution in water, 25% (Merck, catalog number: 1054321000)
Acetonitrile anhydrous, 99.8% (Sigma, catalog number: 271004)
N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide ≥95% containing 1% tert-butyldimethylchlorsilane (MTBSTFA) (Sigma, catalog number: 375934), store at -20°C
Equipment
Cultivation experiments of anaerobic organisms
Scales
Gas piping system
Membrane vacuum pump (Vacuubbrand, catalog number: 19137003)
Vinyl anaerobic chamber (Coy laboratory products) or equivalent
Bottle-top dispensers (Brand, catalog number: 4600360) or equivalent
pH Meter SevenCompact S210 (Mettler Toledo, catalog number: 30130862) or equivalent
Incubator shaker New Brunswick Innova® 44R (Eppendorf, catalog number: M1282-006) or equivalent temperature-controlled shaker
Optical microscope (Zeiss, model: Axiostar) or equivalent
Thoma cell counting chamber (VWR, catalog number: 631-0697)
Harvesting of biomass and preparation of cell extracts
Sorvall centrifuge RC6+ (Thermo Fisher Scientific, catalog number: 46915) or equivalent
Fixed-angle rotor F9-4x1000y (Thermo Fisher Scientific, catalog number: 096-041032)
Table centrifuge 5810/5810 R (Eppendorf, catalog number: 5811000015) or equivalent
Rotor A-4-81 (Eppendorf, catalog number: 5810718007)
Table microcentrifuge 5417R (Eppendorf) or equivalent
Rotor F45-30-11 (Eppendorf)
Electronic pipette (Eppendorf, model: Easypet) or equivalent
Vinyl anaerobic chamber (Coy laboratory products) or equivalent
Optical microscope (Zeiss, model: Axiostar or equivalent)
Ultrasonic homogenizer Sonopuls (BANDELIN electronic, catalog number: 2451) or equivalent
Aluminum cooling block
-80°C Freezer
Enzyme assays
Cary 3500 UV-Vis spectrophotometer (Agilent) or equivalent; please note that working with thermophiles requires a temperature-controlled cuvette holder (air-cooled Peltier temperature control in our case)
Cuvettes, 400 µL, light path 10 mm (Hellma, catalog number: 115-10-40) or equivalent
Syringe Hamilton, 50 µL (Carl Roth, catalog number: X048.1) or equivalent
Rubber stoppers for cuvettes
Vinyl anaerobic chamber (Coy laboratory products) or equivalent
Thermomixer (Eppendorf, model: comfort) or equivalent
Vacuum pump
Ultrasonic bath (BANDELIN electronic, model: DT 100) or equivalent
Ultra-high-performance liquid chromatography instrument (UHPLC) (Agilent, model: 1290 Infinity II LC System) or equivalent
Column for reversed-phase LC separations (phase EC-C18) (Agilent, catalog number: 699675-902) or equivalent
Pipette set (RAININ) or equivalent
Isotopologue profiling (based on GC-MS analysis)
Freeze-dryer Alpha 2-4 LDplus (Martin Christ, Osterode, Germany) or equivalent
Oven (Binder, model E28, Tuttlingen, Germany,) or equivalent
Heating block Techne® DRI Block DB 2A (Merck, Darmstadt, Germany) or equivalent
GCMS-QP 2010 plus with autosampler AOC20i (Shimadzu, Duisburg, Germany) or equivalent
Silica capillary column (equity TM-5; 30 m by 0.25 mm; 0.25 µm film thickness; Sigma-Aldrich, Supelco, catalog-number 28098-U)
Software
Interactive Codon Analysis INCA software (Supek and Vlahovicek, 2005; http://bioinfo.hr/software-tools/)
National Center for Biotechnology Information (NCBI) BLAST (http://www.ncbi.nlm.nih.gov/BLAST/)
Labsolutions (Shimadzu, https://www.shimadzu.de/labsolutions%E2%84%A2-lcms-software-simple-analytical-platform)
Isotopo (Ahmed et al., 2014, https://www.uni-wuerzburg.de/tr34/software-developments/isotopo/)
Microsoft Excel (Microsoft, 2016, https://www.microsoft.com/en-us/microsoft-365/excel)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Steffens, L., Pettinato, E., Steiner, T. M., Eisenreich, W. and Berg, I. A. (2022). Tracking the Reversed Oxidative Tricarboxylic Acid Cycle in Bacteria. Bio-protocol 12(6): e4364. DOI: 10.21769/BioProtoc.4364.
分类
微生物学 > 微生物生物化学 > 蛋白质
生物化学 > 蛋白质 > 活性
生物化学 > 其它化合物
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link