- EN - English
- CN - 中文
TUNEL Labeling to Detect Double-stranded DNA Breaks in Caenorhabditis elegans Gonads
TUNEL 标记检测秀丽隐杆线虫性腺中的双链 DNA 断裂
发布: 2022年03月20日第12卷第6期 DOI: 10.21769/BioProtoc.4351 浏览次数: 2611
评审: Demosthenis ChronisAnonymous reviewer(s)
Abstract
Analysis of DNA double strand breaks (DSBs) is important for understanding dyshomeostasis within the nucleus, impaired DNA repair mechanisms, and cell death. In the C. elegans germline, DSBs are important indicators of all three above-mentioned conditions. Although multiple methods exist to assess apoptosis in the germline of C. elegans, direct assessment of DSBs without the need for a reporter allele or protein-specific antibody is useful. As such, unbiased immunofluorescent approaches can be favorable. This protocol details a method for using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) to assess DNA DSBs in dissected C. elegans germlines. Germlines are co-labeled with DAPI to allow for easy assessment of DNA DSBs. This approach allows for qualitative or quantitative measures of DNA DSBs.
Graphic abstract:
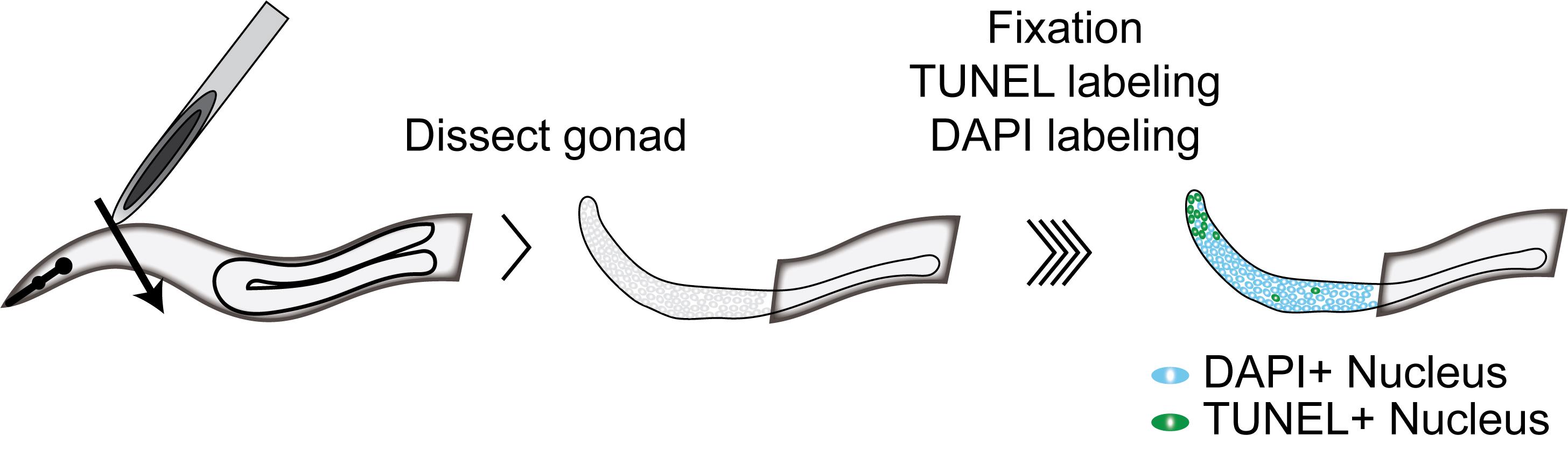
Schematic for TUNEL labeling of C. elegans germlines.
Background
Many stresses, nuclear or otherwise, can result in germline phenotypes in C. elegans, including reduced brood size or complete sterility. When trying to understand the mechanism resulting in germline defects, it is necessary to investigate the possibility of increased DNA double strand breaks that are likely to lead to apoptosis. Although techniques are well established to assess germline apoptosis [e.g., analysis of cell corpses; CED-3::GFP transgenes (Chen et al., 2016)] and DNA damage [e.g., LacZ reporters transgenes (Pontier and Tijsterman, 2009); immunolabeling for DSB repair markers, such as RAD-51 and RPA-1 (Bae et al., 2020; Hinman et al., 2021)] in the C. elegans germline, inclusion of a genetically-encoded reporter can be complicated or impossible in the case of genetic linkage. Therefore, biochemical approaches may offer more flexibility in assessment of DNA DSBs. As presented here, TUNEL labeling can identify DNA DSBs in nearly all C. elegans genetic backgrounds (excluding those without a gonad). This approach not only eliminates the need to generate new mutant strains containing the reporter, but allows for parallel preparation and analysis of multiple samples. This protocol is an adaptation of a protocol used in Parusel (2006) and exactly as described inKropp et al. (2021).
Materials and Reagents
25 × 75 mm Superfrost Plus Slides (Diagger, catalog number: EF15978Z), store at room temperature
25 × 25-1 Microscope Cover Glass (Fisherbrand, catalog number: 12-542-C), store at room temperature
23 G 1¼ Precision Glide Needle (BD Bioscience, catalog number: 305120), store at room temperature
KimWipes (Kimtech Science, Kimberly-Clark Professional, catalog number: 05511), store at room temperature
M9 Buffer (IPM Scientific, catalog number: 11006-517), store at room temperature
10× PBS (KD Medical, catalog number: RGF-3210), store at room temperature
Poly-L-lysine solution (Sigma Life Science, catalog number: P8920-100mL), store at room temperature
Paraformaldehyde Powder (Sigma-Aldrich, catalog number: 158127), store at 4°C
Triton X-100 (Sigma Life Science, catalog number: X100-100mL), store at room temperature
Sodium Citrate tribasic dihydrate powder (Sigma-Aldrich, catalog number: C8532), store at room temperature
TUNEL Assay Kit – FITC (Abcam, catalog number: ab66108), store at 4°C and -20°C
DAPI powder (Sigma-Aldrich, catalog number: D9542), store at 4°C
Vectashield (Vector Laboratories, Inc, catalog number: H-1000), store at 4°C
Immersion oil for microscopy (Cargille, catalog number: 16482), store at room temperature
4% paraformaldehyde (PFA) solution (see Recipes).
PTX solution (see Recipes)
100 mM sodium citrate buffer (see Recipes)
DAPI solution (see Recipes)
TUNEL reaction mix from TUNEL Assay kit (see Recipes)
1 M Sodium citrate (see Recipes)
Equipment
SMZ645 stereomicroscope (Nikon, model: SMZ645-L)
Digital Heat block (VWR Scientific Products, catalog number: 13259-050)
Spinning disk confocal microscope
Nikon Eclipse Ti2-E inverted microscope
Yokagawa CSUZ-1 spinning disk
Photometrics Prime95B camera
Nikon Plan Flour 40×/1.3 oil immersion objective
Software
Elements Advanced Research (Nikon, https://www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-advanced-research)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Kropp, P. A., Rhodehouse, K. and Golden, A. (2022). TUNEL Labeling to Detect Double-stranded DNA Breaks in Caenorhabditis elegans Gonads. Bio-protocol 12(6): e4351. DOI: 10.21769/BioProtoc.4351.
分类
发育生物学 > 细胞信号传导 > 胁迫反应
发育生物学 > 细胞信号传导 > 细胞凋亡
分子生物学 > DNA > DNA 损伤和修复
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link