- EN - English
- CN - 中文
A Novel PCR-Based Methodology for Viral Detection Utilizing Mechanical Homogenization
一种基于聚合酶链反应的机械均匀化病毒检测新方法
发布: 2022年03月05日第12卷第5期 DOI: 10.21769/BioProtoc.4349 浏览次数: 2761
评审: Alessandro DidonnaLucy Xie
Abstract
The impact of viral diseases on human health is becoming increasingly prevalent globally with the burden of disease being shared between resource-rich and poor areas. As seen in the global pandemic caused by SARS-CoV-2, there is a need to establish viral detection techniques applicable to resource-limited areas that provide sensitive and specific testing with a logistically conscious mindset. Herein, we describe a direct-to-PCR technology utilizing mechanical homogenization prior to viral PCR detection, which allows the user to bypass traditional RNA extraction techniques for accurate detection of human coronavirus. This methodology was validated in vitro, utilizing human coronavirus 229E (HCoV-229E), and then clinically, utilizing patient samples to test for SARS-CoV-2 infection. In this manuscript, we describe in detail the protocol utilized to determine the limit of detection for this methodology with in vitro testing of HCoV-229E.
Keywords: Coronavirus (新冠病毒)Background
Polymerase chain reaction (PCR) and reverse transcriptase PCR (RT-qPCR)-based methodologies are the current mainstay of detection for human viral infections (Elnifro et al., 2000). These PCR methodologies often rely on multiple chemical extraction steps, for lysis of the viral particles from the clinical sample, followed by a series of washing steps, which are meant to purify the exposed viral genetic material while removing other macromolecules, such as carbohydrates and proteins, that may impact downstream molecular applications (Elnifro et al., 2000). Once the purified genetic material has been isolated, it is then placed into the PCR mixture for detection of a specific targeted genetic sequence, thereby verifying the presence of the virus. This traditional extraction model for PCR detection of virus is highly sensitive and specific in confirming infection (Elnifro et al., 2000). However, as seen in the global pandemic caused by SARS-CoV-2, the stressed global supply chain could not always ensure availability of the chemical reagents needed for these extractions, driving the need for novel diagnostic techniques (Bustin et al., 2020).
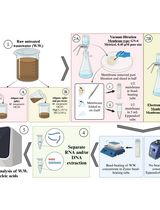
Herein, we describe a methodology where mechanical homogenization was used to lyse viral particles, exposing RNA for accurate detection of the virus when placed directly into PCR, without undergoing further extraction steps or chemical washes (Morehouse et al., 2020). This methodology has been reported utilizing in vitro simulation of clinical viral samples with human coronavirus 229E (HCoV-229E) (Morehouse et al., 2020), as well as in a field validation study with the use of nasopharyngeal and oropharyngeal swabs for the detection of SARS-CoV-2 from patients (Morehouse et al., 2021). This homogenization-based direct-to-PCR technique allows for accurate viral detection of human coronaviruses, without the need for time consuming and costly chemical extraction techniques traditionally used in RT-qPCR detection workflows (Morehouse et al., 2020).
In an effort to improve access to cost effective diagnostics solutions, we demonstrate how human coronavirus can be accurately detected in clinical concentrations with similar sensitivity and specificity to the current gold standard extraction-based techniques (Morehouse et al., 2020 and Morehouse et al., 2021). It is critical that innovative diagnostic solutions such as this are continually developed with concern for the logistical challenges of providing accurate viral detection in resource-limited settings. Through the utilization of mechanical homogenization, we were able to detect HCoV-229E off spiked swabs, simulating clinically relevant concentrations of virus on nasopharyngeal swabs as an initial proof of concept study for this technique (Morehouse et al., 2021). The homogenization-based direct-to-PCR methodology was then further validated with patient samples screening for SARS-CoV-2 in a later study (Morehouse et al., 2021). In this manuscript, we lay out the detailed protocol utilized in the initial proof of concept testing of our homogenization-based direct-to-PCR technology, utilizing simulated nasopharyngeal swabs with spiked HCoV-229E to determine the limit of detection for this proposed methodology.
Materials and Reagents
2 mL screw cap tubes (Omni International Inc., catalog number: 19-628) prefilled with 1 mL of viral transport media (VTM)
Corning 2 mL internal threaded polypropylene cryogenic vials, self-standing with round bottom (Corning, catalog number: 431386)
Cotton tipped swabs (Dynarex, catalog number: 4305)
PCR tubes: 96 well plate format (Bio-Rad, catalog number: HSP9601) with plate sealing film (Bio-Rad, catalog number: MSB1001)
Viral transport media, made following the United States Centers for Disease Control and Prevention (US CDC) protocol (SOP#: DSR-052-05). While we utilized the US CDC VTM made in our laboratory for this protocol, the authors are aware that many other commercially available VTMs are eligible to be substituted based on the availability of reagents in any given laboratory environment.
HCoV-229E virus stock in DMEM, prepared from cell culture
HCoV-229E (ATCC, catalog number: VR-740) cultured following manufacturers recommendations on MRC-5 cells (ATCC, catalog number: CCL-171) in standard DMEM (Gibco, catalog number: 11965092). Cell culture media was collected following manufacturer’s instructions 72 h after inoculation, and then utilized as the virus stock for these experiments.
Corning 75 cm2 U-shaped canted neck cell culture flask with vent cap (Corning, catalog number: 430641U)
Corning PureCoat Amine 6-well tissue culture plates (Corning, catalog number: 354721)
Luna Universal One-Step RT-qPCR Kit (New England Biolabs, catalog number: E3005S)
HCoV-229E N forward and reverse primer set (Integrated DNA Technologies, Custom Oligonucleotide Purchase)
N gene forward primer: 5’-AGGCGCAAGAATTCAGAACCAGAG-3’
N gene reverse primer: 5’-AGCAGGACTCTGATTACGAGAAAG-3’
Molecular grade agarose (Bio-Rad, catalog number: 161-3101)
Ethidium bromide (Bio-Rad, catalog number: 161-0433)
TBE buffer (IBI Scientific, catalog number: IB70153)
100 bp molecular ruler (Bio-Rad, catalog number: 1708202)
Trypan blue (Bio-Rad, catalog number: 1450021)
Equipment
Bead Ruptor Elite (Omni International Inc., catalog number: 19-042E)
CFX connect (Bio-Rad, catalog number: 1855201)
Gel Doc EZ Imaging System (Bio-Rad, catalog number: 1708270)
Rainin Classic manual pipettes (Rainin, catalog number: 17008708)
ThermoFisher Fresco 17 microcentrifuge (Thermo Fisher Scientific, catalog number: 75002421)
Gel electrophoresis chamber (BioRad, catalog number: 1704487EDU)
Thermo Scientific TSX ultra-low freezer (Thermo Fischer Scientific, catalog number: 09313868)
Software
CFX Software Bio-Rad CFX Maestro 1.1 Version 4.1.2433.1219
EZ Imager Software Image LabTM Version 6.0.0 build 25
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Morehouse, Z. P., Proctor, C. M., Ryan, G. L. and Nash, R. J. (2022). A Novel PCR-Based Methodology for Viral Detection Utilizing Mechanical Homogenization. Bio-protocol 12(5): e4349. DOI: 10.21769/BioProtoc.4349.
分类
微生物学 > 病原体检测 > PCR
分子生物学 > RNA > qRT-PCR
分子生物学 > RNA > RNA 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link