- EN - English
- CN - 中文
Immunoprecipation Assay to Quantify the Amount of tRNAs associated with Their Interacting Proteins in Tissue and Cell Culture
免疫沉淀法定量组织和细胞培养中与其相互作用蛋白相关的tRNAs的数量
发布: 2022年02月20日第12卷第4期 DOI: 10.21769/BioProtoc.4335 浏览次数: 3576
评审: Pilar Villacampa AlcubierreDaniel Louis KissThirupugal Govindarajan
Abstract
Transfer RNAs (tRNAs) are highly abundant species and, along their biosynthetic and functional path, they establish interactions with a plethora of proteins. The high number of nucleobase modifications in tRNAs renders conventional RNA quantification approaches unsuitable to study protein-tRNA interactions and their associated functional roles in the cell. We present an immunoprecipitation-based approach to quantify tRNA bound to its interacting protein partner(s). The tRNA-protein complexes are immunoprecipitated from cells or tissues and tRNAs are identified by northern blot and quantified by tRNA-specific fluorescent labeling. The tRNA interacting protein is quantified by an automated western blot and the tRNA amount is presented per unit of the interacting protein. We tested the approach to quantify tRNAGly associated with mutant glycyl-tRNA-synthetase implicated in Charcot-Marie-Tooth disease. This simple and versatile protocol can be easily adapted to any other tRNA binding proteins.
Graphic abstract:
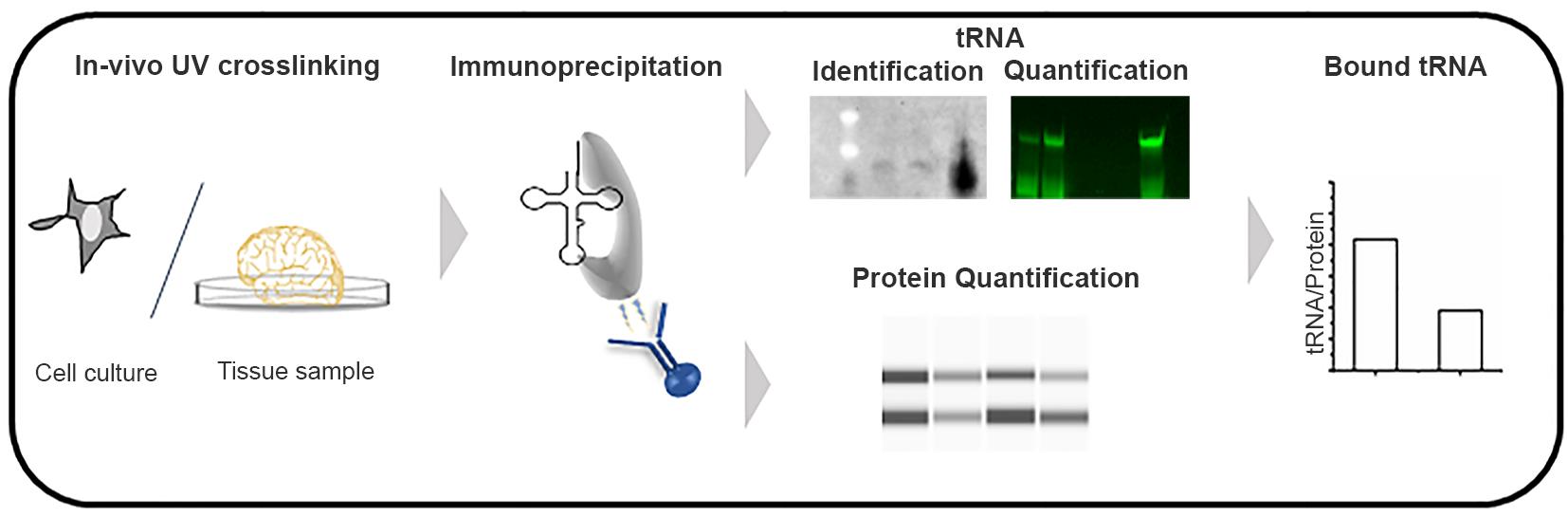
Figure 1. Schematic of the tRNA-Immunoprecipitation approach.
Background
tRNAs are ubiquitous molecules, representing 4–10% of all cellular RNAs. tRNAs undergo complex biogenesis during which they interact with different protein entities, including tRNA-splicing proteins, tRNA-base modifying enzymes, tRNA-charging enzymes, 3’-end modification and repair enzymes, and various nucleases, thereby generating active tRNA fragments or completely degrading tRNAs (Betat and Mörl, 2015; Kirchner and Ignatova, 2015; Fernández-Millán et al., 2016; Barciszewska et al., 2016; Schmidt and Matera, 2020; Tosar and Cayota, 2020). tRNAs are crucial components of the translation machinery and are charged at the 3’ ends with their cognate amino acid by an aminoacyl-tRNA-synthetase. Mutations in tRNAs or genes encoding tRNA-interacting partners are linked to complex human pathologies, with intricate heterogeneity at both the cell and tissue levels that modulates the disease (Abbott et al, 2014). Thus, a method for quantitative detection of tRNA-binding-protein interactions that can be widely used to study disease-related alterations of tRNA interactome in living cells and tissues is an urgent need.
Traditional methods to detect RNA-protein interactions include RNA immunoprecipitation (RIP) and crosslinking and immunoprecipitation (CLIP), both of which use antibodies to immunoprecipitate RNA-protein complexes, followed by identification of RNAs by sequencing. Unlike mRNA sequencing, despite recent advances (Zheng et al., 2015; Behrens et al., 2021) tRNA sequencing still has limited quantitative resolution towards many tRNA isoacceptors, likely because of their complex modification pattern (Orioli, 2017; Kimura et al., 2020; Warren et al., 2021). Combining immunoprecipitation (IP) of the RNA-protein complexes with tRNA-tailored detection (Figure 1), we have developed a new twist of the classic IP method that is suitable for quantifying tRNA-protein interactions in living cells. In a recent study, we have used this approach to quantify alterations in tRNA binding to mutated glycyl-tRNA-synthetase (GlyRS), which has been implicated in Charcot-Marie-Tooth (CMT) disease (Zuko et al., 2021). In a CMT-mouse model GarsC201R/+, we observed a stronger association of tRNAsGly with the mutant GlyRS, thus depleting the glycyl-tRNAGly pool and causing ribosome stalling at Gly codons (Zuko et al., 2021). The tRNA-IP methodology identifies and quantifies tRNAs bound to GlyRS in native conditions in tissues. This protocol is easily adapted to other aminoacyl-tRNA-synthetases or any tRNA-binding proteins, to quantify interactions in native conditions, in both cell culture and tissue.
Materials and Reagents
Pipette tips (filtered) (Sarstedt)
Syringe 1 mL with 26 G needles (BD Plastipak)
Cell Scraper, DNase/RNase free (Techno Plastic Products)
RNase-free 1.5 mL microtubes (Carl Roth, catalog number: CNT2.1)
RNase-free 1.5 mL amber microtubes (Carl Roth, catalog number: ENH0.2)
15 cm cell culture dishes (Sarstedt, catalog number: 83.3903)
3.5 cm cell culture dishes (Sarstedt, catalog number: 82.1135.500)
GarsC201R/+ mice (C3H.C-GarsC201R/H) (Achilli et al., 2009)
293 (HEK293) cell line 293 (HEK293) (ATCC CRL-1573TM)
Gibco Dulbecco's modified Eagle medium (DMEM) (Pan Biotech, catalog number: P04-03500)
Fetal bovine serum (FBS) (Pan Biotech, catalog number: P30-3302)
L-glutamine (Serva, catalog numbers: 22942)
PBS (GibcoTM, catalog number: 70011044)
0.25% Trypsin-EDTA (Pan Biotech, catalog number: P10-019500)
Triton® X-100 (Sigma, catalog number: T9284)
Tris Base (Sigma, catalog number: T6066)
cOmplete TM, EDTA-free protease inhibitor cocktail (Roche, catalog number: 11873580001)
Urea (Carl Roth, catalog number: 57-13-6)
NaCl (Carl Roth, catalog number: HN00.2)
Acid-phenol:chloroform, pH 4.5 (Invitrogen, catalog number:AM9720)
3 M sodium acetate, pH 5.2 (Carl Roth, catalog number: 6773.2)
Glycogen (Invitrogen, catalog number: AM9510)
Absolute ethanol (Carl Roth, catalog number: 9065.4)
Isopropanol (Carl Roth, catalog number: 7343.2)
40% polyacrylamide (Carl Roth,catalog number: A516.1 )
RNase inhibitor SUPERase-In (Invitrogen, catalog number: AM2694)
NP-40 (Sigma, catalog number: 6507)
DTT (Carl Roth, catalog number: 6908.2)
SDS (Carl Roth, catalog number: CN30.3)
Sodium deoxycholate detergent (SDC) (Thermo, catalog number: 89904)
EDTA (AppliChem, catalog number: A5097,1000)
BSA (Carl Roth, catalog number: T844.3)
Trisodium citrate (Carl Roth, catalog number: 3580.1)
Hybond-N membrane (GE Healthcare Life Sciences, RPN2250B)
T4 DNA ligase (NEB, catalog number: M0202)
DMSO (Sigma, catalog number: D8418)
Salmon sperm DNA (Invitrogen, catalog number: 15632011)
Anti-GARS antibodies (Abcam, catalog number: ab42905; Proteintech, catalog number: 15831-AP)
Pierce protein G magnetic beads (Thermo, catalog number: 88847)
RevertAid H Minus reverse transcriptase (Thermo, catalog number: K1631)
10 mM DNTPs (Thermo, catalog number: R0181)
T7 RNA polymerase (Thermo, catalog number: EP0113)
NTPs (Thermo, catalog number: R0481)
Crush and soak buffer (see Recipes)
2× RNA loading formamide dye (see Recipes)
20× saline sodium citrate buffer (SSC buffer; see Recipes)
Cell lysis buffer (see Recipes)
Tissue lysis buffer (see Recipes)
Wash buffer for cell culture (see Recipes)
Wash buffer for tissue (see Recipes)
1× SDS buffer (see Recipes)
Hybridization buffer (see Recipes)
Note: Except for the anti-GARS antibodies, all other reagents can be purchased from any other supplier.
Equipment
Standard molecular biology equipment
UV crosslinker (Analytik Jena, catalog number: CL-1000)
CellCrusher tissue pulverizer (Kisker, catalog number: 538004)
Eppendorf tube rotator (Eppendorf, catalog number: R5010)
Spectrophotometer UV-vis (DeNovix DS-C)
ChemiDocTM MP Imaging System multiplex fluorescence, chemiluminescence (Bio-Rad)
Magnetic bead separator (Thermo Fisher, catalog number: CS15000)
Jess automated western blots system (Protein Simple)
Software
Fiji ImageJ (ImageJ, https://imagej.net/software/fiji/)
Compass software for simple western (Protein Simple, a biotechne brand, https://www.proteinsimple.com/software_compass_simplewestern.html)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Das, S., Zuko, A., Thompson, R., Storkebaum, E. and Ignatova, Z. (2022). Immunoprecipation Assay to Quantify the Amount of tRNAs associated with Their Interacting Proteins in Tissue and Cell Culture. Bio-protocol 12(4): e4335. DOI: 10.21769/BioProtoc.4335.
- Zuko, A., Mallik, M., Thompson, R., Spaulding, E. L., Wienand, A. R., Been, M., Tadenev, A. L. D., van Bakel, N., Sijlmans, C., Santos, L. A., et al. (2021). tRNA overexpression rescues peripheral neuropathy caused by mutations in tRNA synthetase. Science 373(6559): 1161-1166.
分类
神经科学 > 神经系统疾病 > 神经退行性病变
生物化学 > RNA
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link