- EN - English
- CN - 中文
A Cell-based Screening Method Using an Intracellular Antibody for Discovering Small Molecules Targeting Hard-to-drug Proteins
一种基于细胞的筛选方法,使用细胞内抗体发现靶向困难药物蛋白质的小分子
发布: 2022年02月20日第12卷第4期 DOI: 10.21769/BioProtoc.4324 浏览次数: 3895
评审: Alessandro DidonnaYoshihiro AdachiAnonymous reviewer(s)
Abstract
Targeting hard-to-drug proteins, such as proteins functioning by protein-protein interactions (PPIs) with small molecules, is difficult because of the lack of well-defined pockets. Fragment or computational-based methods are usually employed for the discovery of such compounds, but no generic method is available to quickly identify small molecules interfering with PPIs. Here, we provide a protocol describing a generic method to discover small molecules inhibiting the interaction between an intracellular antibody and its target, in particular for proteins that are hard to make in recombinant form. This protocol reports a versatile and generic method that can be applied to any target/intracellular antibody. Because it is a cell-based assay, it identifies chemical matters that are already displaying advantageous cell permeability properties.
Graphic abstract:
Cell-based intracellular antibody-guided small molecule screening.
Background
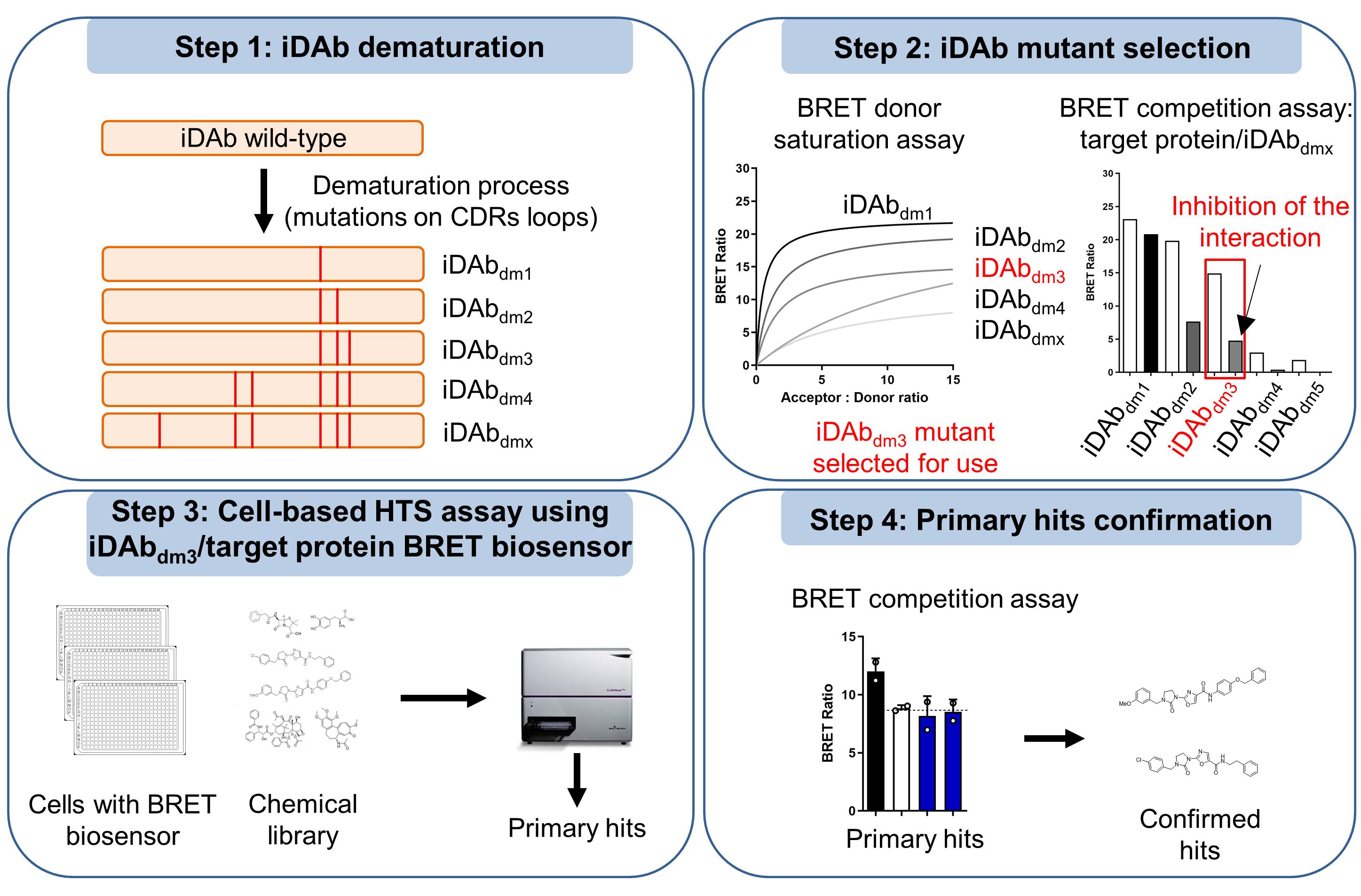
Targeting protein-protein interactions (PPIs) with small molecules is a challenging task, due to their relatively large interaction interface and the lack of well-defined pockets. Hence, implementing generic methods to discover such compounds would be highly valuable. Interestingly, PPIs can be inhibited by intracellular macromolecules, such as intracellular single domain antibodies (hereafter named iDAbs) (Tanaka et al., 2007 and 2011), or antibody mimetics (Guillard et al., 2017; Bery et al., 2019b). The advantages of such reagents are their high specificity for their target and their ability to discriminate isoforms of protein families (Bery et al., 2019a and 2019b), which can enable the identification of novel targetable regions. Therefore, we developed an intracellular antibody-guided method to screen small molecules named antibody-derived compound (Abd). We first used this method in a competitive surface plasmon resonance (cSPR) set-up to identify anti-RAS small molecules that bind at the combining site of an anti-RAS iDAb (Quevedo et al., 2018). However, favourable binding properties of the intracellular antibody with its target (very high affinity, high Kon, and low Koff) and recombinant protein targets that are easily expressed are required for this biochemical Abd assay. Furthermore, the selected compounds should have advantageous properties in cellular uptake with this assay. Therefore, new versatile methods would be important to rapidly discover small molecules against difficult drug targets.
Accordingly, we implemented an Abd cell-based screening method. We developed bioluminescence resonance energy transfer 2 (BRET2)-based LIM domain only 2 (LMO2) biosensors, based on the strategy of RAS biosensors (Bery et al., 2018), to monitor the interaction between the chromosomal translocation protein LMO2 and its iDAb. This technique enabled the selection of compounds that interfere with this interaction in cells (Bery et al., 2021). The cell-based Abd assay is a versatile method, as the measured in vitro affinity of the iDAbs for their target is not a limitation. High-affinity iDAbs could be dematured to reduce their binding for use in the cell-based assay (Tanaka et al., 2021). In addition, this method is not limited by proteins that are difficult to express and/or to purify in recombinant form. Finally, the intrinsic advantage of cell-based assays is that the compounds already have the characteristic of cell entry, a highly relevant property for the use of small molecules as drugs.
This protocol could be implemented in any intracellular antibody (or antibody mimetic) targeting challenging proteins, such as transcription factors or chromosomal translocation proteins.
Materials and Reagents
6-well plates (Corning, catalog number: 3516)
Corning HYPERFlask M cell culture vessels (Corning, catalog number: 10030)
Corning® 250 mL PP centrifuge tubes (Corning, catalog number: 430776)
White 96-well plates, clear bottom (PerkinElmer, catalog number: 6005181)
White 384-well plates, clear bottom (PerkinElmer, catalog number: 6007480)
Aluminum foil
White adhesive bottom seal for 96 and 384-well plates (PerkinElmer, catalog number: 6005199)
HEK293T cells (ATCC# CRL-3216, RRID:CVCL_0063)
Chemically competent DH5α cells (New England Biolabs (NEB), catalog number: C2987H)
Penicillin/streptomycin (PS) (Thermo Fisher, catalog number: 10378016)
Trypsin (Thermo Fisher, catalog number: 25300054)
DMEM (Thermo Fisher, catalog number: 10566016)
OptiMEM medium (Thermo Fisher, catalog number: 31985070)
Lipofectamine 2000 (Thermo Fisher, catalog number: 11668019)
Fetal bovine serum (FBS; Sigma, catalog number: F2442)
Phosphate-buffered saline (PBS; Thermo Fisher, catalog number: 10010023)
RLuc8 construct (see Table 1, sequence freely available in Bery et al., 2018)
GFP2 constructs (see Table 1, sequences freely available in Bery et al., 2021)
LMO2 competitors’ plasmids (see Table 1, sequences freely available in Bery et al., 2018)
OptiMEM no red phenol (Thermo Fisher, catalog number: 11058021)
Coelenterazine 400a (Cayman Chemicals, catalog number: 16157). Store at -20°C if not reconstituted.
100% ethanol (Sigma, catalog number: 32221-M54)
Dimethyl Sulfoxide (DMSO; Sigma, catalog number: D2650)
QIAquick Gel Extraction Kit (QIAGEN, catalog number: 28704)
Agarose (Sigma, catalog number: A9539-500G)
Restriction enzymes: NotI-HF, XbaI, PspOMI (NEB, catalog numbers: R3189S, R0145S and R0653S respectively)
Vent DNA polymerase (NEB, catalog number: M0254S)
T4 DNA Ligase (NEB, catalog number: M0202S)
Ampicillin (Sigma, catalog number: A0166)
pEF-myc-cyto, empty plasmid (Invitrogen, catalog number: V89120)
In-house library of 10,720 compounds (comprising 6991 compounds from BioFocus and 3729 from ChemBridge). Stored at -20°C
BRET medium (see Recipes)
BRET substrate (see Recipes)
Table 1. List of donor and acceptors constructs available for BRET2-based LMO2 biosensors
RLuc8 Donor construct GFP2 Acceptors constructs Competitors constructs LMO2 iDAb LMO2 WT iDAb LMO2-myc (positive control, Bery et al., 2018) iDAb LMO2 S55A/T107A
(iDAb LMO2dm)iDAb RAS-myc (negative control, Bery et al., 2018) iDAb LMO2 S28G/H31G/S55A/T107A
(iDAb LMO2dm1)iDAb LMO2 S55A/E102A/T107A
(iDAb LMO2dm2)iDAb LMO2 S28G/H31G/S55A/E102A/T107A
(iDAb LMO2dm3)iDAb LMO2 S28G/H31G/S55A/S103A/T107A
(iDAb LMO2dm4)iDAb LMO2 S55A/E102A/S103A/T107A
(iDAb LMO2dm5)iDAb LMO2
S28G/H31G/S55A/E102A/S103A/T107A
(iDAb LMO2dm6)
Equipment
Haemocytometer (Sigma, catalog number: Z359629)
SpeedVac instrument (any supplier)
Janus liquid handling workstation (PerkinElmer)
Echo Acoustic Dispenser (Labcyte)
CLARIOstar microplate reader including the luminescence module (BMG Labtech)
PHERAstar FSX microplate reader with the BRET2 optic filters (BMG Labtech)
Cell culture 37°C incubator (any supplier)
Centrifuge (any supplier)
Software
MARS Data analysis (BMG Labtech, included with the microplate readers)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Bery, N. and Rabbitts, T. H. (2022). A Cell-based Screening Method Using an Intracellular Antibody for Discovering Small Molecules Targeting Hard-to-drug Proteins. Bio-protocol 12(4): e4324. DOI: 10.21769/BioProtoc.4324.
- Bery, N., Bataille, C. J. R., Russell, A., Hayes, A., Raynaud, F., Milhas, S., Anand, S., Tulmin, H., Miller, A. and Rabbitts, T. H. (2021). A cell-based screening method using an intracellular antibody for discovering small molecules targeting the translocation protein LMO2. Sci Adv 7(15): eabg1950.
分类
癌症生物学 > 癌症生物化学
药物发现 > 药物设计
细胞生物学 > 基于细胞的分析方法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link









