- EN - English
- CN - 中文
Optimization of Extracellular Flux Assay to Measure Respiration of Anchorage-independent Tumor Cell Spheroids
优化细胞外通量测定以测量不依赖锚定的肿瘤细胞球体的呼吸
发布: 2022年02月20日第12卷第4期 DOI: 10.21769/BioProtoc.4321 浏览次数: 4568
评审: Masahiro MoritaKimberly Dunham-SnaryAnonymous reviewer(s)
Abstract
Three-dimensional (3D) cell culture models are widely used in tumor studies to more accurately reflect cell-cell interactions and tumor growth conditions in vivo. 3D anchorage-independent spheroids derived by culturing cells in ultra-low attachment (ULA) conditions is particularly relevant to ovarian cancer, as such cell clusters are often observed in malignant ascites of late-stage ovarian cancer patients. We and others have found that cells derived from anchorage-independent spheroids vary widely in gene expression profiles, proliferative state, and metabolism compared to cells maintained under attached culture conditions. This includes changes in mitochondrial function, which is most commonly assessed in cultured live cells by measuring oxygen consumption in extracellular flux assays. To measure mitochondrial function in anchorage-independent multicellular aggregates, we have adapted the Agilent Seahorse extracellular flux assay to optimize measurements of oxygen consumption and extracellular acidification of ovarian cancer cell spheroids generated by culture in ULA plates. This protocol includes: (i) Methods for culturing tumor cells as uniform anchorage-independent spheroids; (ii) Optimization for the transfer of spheroids to the Agilent Seahorse cell culture plates; (iii) Adaptations of the mitochondrial and glycolysis stress tests for spheroid extracellular flux analysis; and (iv) Suggestions for optimization of cell numbers, spheroid size, and normalization of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) values. Using this method, we have found that ovarian cancer cells cultured as anchorage-independent spheroids display altered mitochondrial function compared to monolayer cultures attached to plastic dishes. This method allows for the assessment of mitochondrial function in a more relevant patho/physiological culture condition and can be adapted to evaluate mitochondrial function of various cell types that are able to aggregate into multicellular clusters in anchorage-independence.
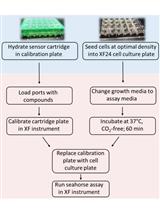
Graphic abstract:
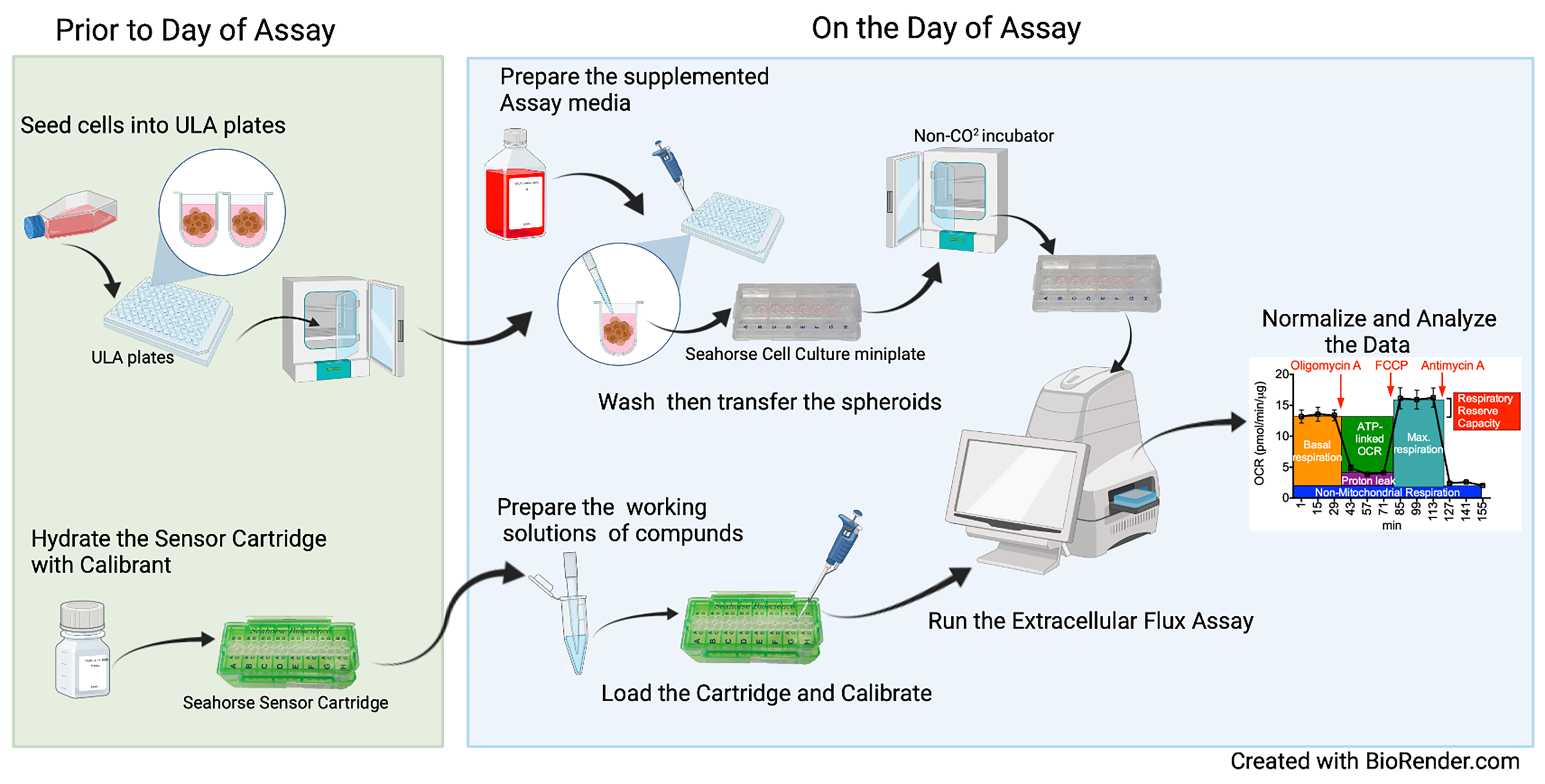
Workflow of the Extracellular Flux Assay to Measure Respiration of Anchorage-independent Tumor Cell Spheroids.
Background
Epithelial ovarian cancer remains the deadliest gynecological malignancy with an estimated five-year survival rate below 29% for patients with advanced stage disease, which is when most patients are first diagnosed (Torre et al., 2018). At these later stages, ovarian cancer cells have metastasized to distant sites across the peritoneum. This spread of ovarian cancer across the peritoneal cavity is facilitated by the passive dissemination of tumor cells directly into the peritoneal ascites fluid. In the malignant ascites, tumor cells can be found as both single cells and as multicellular clusters, also commonly referred to as “spheroids”. Formation of spheroid aggregates, which are enriched in cancer stem cells (Liao et al., 2014; Kenda Suster and Virant-Klun, 2019), are key to the progression of ovarian cancer, as these ensure maintenance of intercellular survival signaling among cells detached from the primary tumor (Tan et al., 2006; Al Habyan et al. 2018). In addition, cancer stem-like cells in spheroids display metabolic flexibility. They shift their metabolism based on nutrient availability and metabolic demands, potentially allowing tumor cells to survive in ascites and invade the neighboring peritoneal organs (Anderson et al.., 2013; Kim et al.., 2020; Ghoneum et al., 2020). To advance our understanding of ovarian cancer metastatic progression and associated changes in tumor metabolism, it is important to develop methods that allow us to study the metabolic changes in anchorage-independent tumor spheroids, which mimic tumor cell clusters found in malignant ascites of patients.
The following protocol utilizes ultra-low attachment (ULA) plates to generate multi-cellular tumor cell spheroid clusters in order to study their metabolic phenotype using the Seahorse XFp (Agilent) extracellular flux assay platform. Spontaneous clustering of tumor cells in anchorage independence is facilitated by the lack of ability to attach to the hydrophilic neutrally charged hydrogel coating of ULA plates. Compared to cells cultured in conventional attached monolayer conditions on plastic surfaces, spheroids more closely reflect the metastatic tumor cells found in anchorage-independent multicellular clusters in malignant ascites. This makes spheroids a good in vitro model to better simulate the in vivo behavior of tumor cells and to study their unique characteristics related to cell survival, drug sensitivity, hypoxia, and tumor metabolism. During optimization of this protocol, we determined that uniform spheroid formation is integral to minimizing variability in extracellular flux measurements. The use of ULA platforms with small well sizes, such as 96 or 384 wells per plate, permits the generation of uniformly sized spheroids by controlling the seeding density. The use of bioprinting in combination with magnetic nanoparticles has been investigated as an alternate method for metabolic studies of 3D tumor-fibroblast co-cultures (Noel et al., 2017). However, for cell types that spontaneously cluster into spheroids in response to anchorage independence, the ULA culture platform represents a relatively straightforward approach to generate uniform multicellular clusters, without the need for further manipulation.
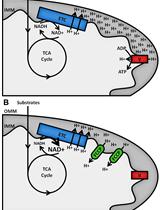
The Seahorse extracellular flux analyzers specifically measure oxygen consumption (OCR) and extracellular acidification rates (ECAR), allowing for the quantification of mitochondrial respiration and an indirect indication of glycolysis, respectively. Several past studies have interrogated mitochondrial function in 3D cultures using single well assays and measured the response to single or a limited number of metabolic stress compounds, with the need to independently determine the rates of oxygen consumption and extracellular acidification (Rodríguez-Enríquez et al., 2008; Schafer et al., 2009; Mandujano-Tinoco et al., 2013; Bloch et al., 2014; Marin-Hernandez et al., 2017). We have optimized the transfer of spheroids from 96 well ULA plates to the Seahorse cell culture plates and the use of this platform for assessment of spheroid bioenergetics, allowing for multi-well analysis and sequential addition of pharmacological agents to manipulate OCR/ECAR. Below are described the use of the mitochondrial stress test and the glycolysis stress test, together with how spheroid culturing and transfer is best achieved. Using substrate limited media and pharmacological approaches, the assay is easily modified to gain further insight into the specific metabolic activity of spheroids. Although not discussed in detail below, iterations of the extracellular flux assay include the palmitate oxidation stress test, which is a modified assay that determines the capacity of cells to oxidize palmitate in the absence or limitation of other exogenous carbon substrates. We refer the reader to the Seahorse Agilent website for alternate assays to assess substrate utilizations. The protocol was developed using the Agilent Seahorse XFp Metabolic Analyzer, and can be applied to the Seahorse XF96 format, as these two platforms utilize cell culture plates with similar surface areas and media volumes. It can also be adapted for the larger surface area of the XF24 platform wells. Hence, the method explained in this paper can be used as a master template for performing not only the mentioned mitochondrial and glycolysis stress tests, but also altered extracellular flux assays with 3D anchorage-independent tumor cell spheroids.
The method outlined below was originally designed for the study of ovarian cancer spheroids cultured in anchorage-independent conditions, which mimic tumor cell clusters commonly found in malignant ascites of ovarian cancer patient (Kim et al., 2020). It can be adapted to other cancer cell lines, primary cells, stem cell cultures, or cell types where 3D cell culture represents a more patho/physiologically relevant culture condition.
Materials and Reagents
96 Well round bottom ultra-low attachment (ULA) plates (Corning, catalog number: 7007)
0.2 µm sterile syringe filter (polyethersulfone membrane, VWR, catalog number: 28145-501)
T-75 tissue culture flask, vent cap, sterile (Celltreat, catalog number: 229341)
0.2 µm sterile vacuum filtration system with polyethersulfone membrane for cell culture media (VWR, catalog number: 10040-436)
Tumor cells:
ES-2 (ATCC, catalog number: CRL-1978)
NIHOVCAR3 (ATCC, catalog number: HTB-161)
OVCA433 cells (generously provided by Dr. Susan Murphy, Duke University)
Note: Experiments should be limited to cells in culture for less than 10 passages.
Cell culture media:
ES-2 cells: McCoy’s 5A + L-glutamine (Corning, catalog number: 10-050-CV), supplemented with 10% FBS
OVCAR3 cells: RPMI 1640 + L-glutamine (Gibco, catalog number: 11875119), supplemented with 20% FBS and 0.01 mg/mL bovine insulin
OVCA433 cells: RPMI 1640 + L-glutamine (Corning, catalog number: 10-040-CV) supplemented with 10% FBS
Note: Media are sterile filtered using the following 0.2 µm vacuum Filters. If desired, Penicillin/Streptomycin may be added to the culture media, depending on cell lines used.
Fetal Bovine Serum (FBS) (VWR, catalog number: 97068-085)
Phosphate buffered saline (PBS, sterile; Corning, catalog number: 21-040-CV)
0.25% Trypsin-EDTA (sterile) (Gibco, catalog number: 25200114)
Seahorse XFp FluxPaks includes Seahorse XFp cell culture miniplates (8 wells), XFp sensor cartridges (8 wells) and XF calibrant solution (Agilent, catalog number: 103022-100)
Seahorse XF base assay media (DMEM, Agilent, catalog number: 103575-100; RPMI, Agilent, catalog number: 103576-100), supplemented on the day of assay as described below, pH 7.4
XF 100 mM Pyruvate Solution (Agilent, catalog number: 103578-100); alternatively sterile 100 mM Pyruvate solution may be sought from other vendors or made in house.
XF 200 mM Glutamine Solution (Agilent, catalog number: 103579-100), alternatively sterile 200 mM Glutamine solution may be sought from other vendors or made in house.
Sterile DEPC-treated ddH2O (Quality Biological, catalog number: 351-068-131); alternatively sterile autoclaved ddH2O can be used.
PierceTM BCA Protein Assay (Thermo Scientific, catalog number: 1859078)
Sodium chloride
Tris-HCl pH 8.0 (Tris Base; Sigma, catalog number: TRIS-RO)
NP40/IgepalCA-630 (Sigma, catalog number: 13021)
Sodium deoxycholate (Sigma, catalog number: D6750)
Sodium phosphate dibasic (Sigma, catalog number: S9763)
Sodium phosphate monobasic (anhydrous; Sigma, catalog number: S3139)
Sodium phosphate buffer
Sodium dodecyl sulfate (SDS; Sigma, catalog number: L3771)
Ethylenedinitrilotetraacetic acid (EDTA; Sigma, catalog number: E9884)
Sodium fluoride (NaF; Sigma, catalog number: 201154)
Protease inhibitor cocktail (Thermo Scientific, catalog number: 87785)
D-glucose (Sigma, catalog number: G8270)
2-Deoxy-D-glucose, 2-DG (Sigma, catalog number: D8375)
Oligomycin A (Sigma, catalog number: 75351) (see Recipes, Table 3)
FCCP (Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; Sigma, catalog number: C2920) (see Recipes, Table 3)
Antimycin A (Sigma, catalog number: A8674) (see Recipes, Table 3)
Rotenone (Sigma, catalog number: 45656) (see Recipes, Table 3)
RIPA lysis buffer (see Recipes)
Equipment
Agilent Seahorse XFp extracellular flux analyzer (Agilent, model: S7802A)
Humidified cell culture incubator (37°C, 5% CO2)
Non-CO2 Humidified cell culture incubator (37°C)
Tissue culture tabletop centrifuge compatible for 15 mL Falcon tubes (Eppendorf, model: Centrifuge 5702)
Tissue culture light microscope with 4× objective and camera
Laminar flow biosafety cabinet (Thermo Scientific, model: 1300 Series A2)
Software
Seahorse Wave Desktop 2.6 Software (Agilent, https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolic-analysis/xf-software/seahorse-wave-desktop-software-740897)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Javed, Z., Worley, B. L., Stump, C., Shimko, S. S., Crawford, L. C., Mythreye, K. and Hempel, N. (2022). Optimization of Extracellular Flux Assay to Measure Respiration of Anchorage-independent Tumor Cell Spheroids. Bio-protocol 12(4): e4321. DOI: 10.21769/BioProtoc.4321.
分类
癌症生物学 > 细胞能量学 > 细胞生物学试验 > 新陈代谢
细胞生物学 > 细胞新陈代谢 > 呼吸测量法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link