- EN - English
- CN - 中文
Listeria innocua Biofilm Assay Using NanoLuc Luciferase
用NanoLuc荧光素酶检测李斯特菌生物膜
发布: 2022年02月05日第12卷第3期 DOI: 10.21769/BioProtoc.4308 浏览次数: 3376
评审: Alba BlesaVinai Chittezham ThomasTimo A Lehti
Abstract
Biofilms serve as a bacterial survival strategy, allowing bacteria to persist under adverse environmental conditions. The non-pathogenic Listeria innocua is used as a surrogate organism for the foodborne pathogen Listeria monocytogenes, because they share genetic and physiological similarities and can be used in a Biosafety Level 1 laboratory. Several methods are used to evaluate biofilms, including different approaches to determine biofilm biomass or culturability, viability, metabolic activity, or other microbial community properties. Routinely used methods for biofilm assay include the classical culture-based plate counting method, biomass staining methods (e.g., crystal violet and safranin red), DNA staining methods (e.g., Syto 9), methods that use metabolic substrates to detect live bacteria (e.g., tetrazolium salts or resazurin), and PCR-based methods to quantify bacterial DNA. The NanoLuc (Nluc) luciferase biofilm assay is a viable alternative or complement to existing methods. Functional Nluc was expressed in L. innocua using the nisin-inducible expression system and bacterial detection was performed using furimazine as substrate. Concentration dependent bioluminescence signals were obtained over a concentration range greater than three log units. The Nluc bioluminescence method allows absolute quantification of bacterial cells, has high sensitivity, broad range, good day-to-day repeatability, and good precision with acceptable accuracy. The advantages of Nluc bioluminescence also include direct detection, absolute cell quantification, and rapid execution.
Graphic abstract:
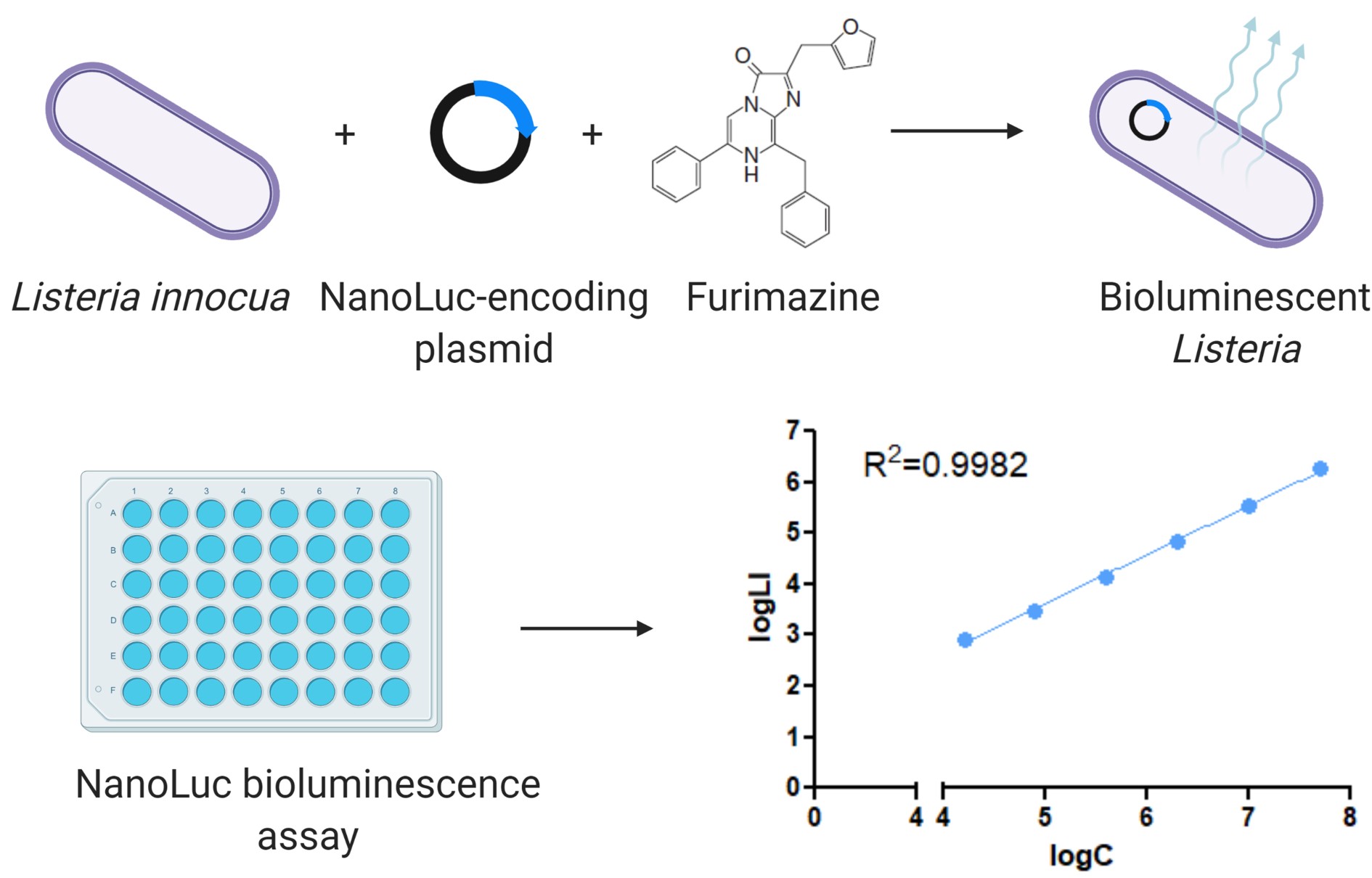
Engineering Listeria innocua to express NanoLuc and its application in bioluminescence assay.
Background
Bacteria form biofilms as a survival strategy, as they enable bacteria to survive and persist in adverse environmental conditions. Within biofilms, bacteria are protected from the human immune system and a variety of environmental factors (e.g., UV, acids, desiccation, salinity, antimicrobials, disinfectants). Opportunistic pathogenic bacteria commonly associated with food poisoning are linked to biofilms formed on surfaces in the food supply chain, and are a significant public health concern. These biofilms can cause persistent contamination and recurrent infections (Donlan, 2002; Chlebicz and Slizewska, 2018; Flemming and Wuertz, 2019). Listeria monocytogenes, the causative agent of listeriosis, is transmitted through the consumption of contaminated food and is associated with high hospitalization and mortality rates compared to other foodborne infectious diseases (EFSA and ECDC, 2019). L. monocytogenes can successfully colonize food processing environments, where it can persist in biofilms and cause outbreaks over several years (Buchanan et al., 2017). The non-pathogenic Listeria innocua has been proposed as a surrogate organism for L. monocytogenes (Costa et al., 2018). Their genomes are conserved (more than 2,500 orthologous genes) and co-linear (Glaser et al., 2001), and they share physiological similarities. However, L. innocua lacks the virulence factors of L. monocytogenes (Fairchild and Foegeding, 1993; Friedly et al., 2008; Silva-Angulo et al., 2015; Costa et al., 2018), so it can be used in Biosafety Level 1 laboratories. Biofilms formed by different strains of L. monocytogenes and L. innocua have often been described as thin and structurally variable (Costa et al., 2018; Lee et al., 2019). Currently available methods target thick and multilayer biofilms, far from the thin biofilms of Listeria.
Several methods have been developed for the quantification of cells and evaluation of listerial biofilms, including different approaches to determine biofilm biomass or culturability, viability, metabolic activity, or other cell properties. These include the classical culture-based plate counting method, biomass staining methods (e.g., crystal violet and safranin red), DNA staining methods (e.g., Syto 9), the use of chromogenic or fluorogenic metabolic substrates to detect live bacteria (e.g., tetrazolium salts or resazurin), and qPCR and digital droplet PCR to quantify bacterial DNA (Stiefel et al., 2016; Klančnik et al., 2017; Ripolles-Avila et al., 2019; Grudlewska-Buda et al., 2020).
The luciferase from Oplophorus gracilirostris, known as NanoLuc (Nluc), has been reported to have excellent properties, including small size (19 kDa), high stability, and high bioluminescence efficiency (150-fold higher compared to other luciferases) (England et al., 2016). The NanoLuc luciferase biofilm assay was compared with the other commonly used methods for detecting biofilms in microtiter plates, i.e., conventional plating and CFU counting, crystal violet staining assay, and resazurin fluorescence assay (Berlec et al., 2021). These methods differ in terms of research equipment and chemicals required, time needed to perform the assay, sensitivity and suitability for observing dynamic processes, and biofilm components detected. The Nluc bioluminescence assay detects all phenotypic subpopulations within the growing biofilm, culturable, viable-but-not culturable forms, and also damaged but non-lysed cells. This can be beneficial in assessing the overall biofilm population, which includes slow growing tolerant and persistent cells that are critical for biofilm persistence (Matereke and Okoh, 2020). Crystal violet staining binds to negatively charged molecules in the biofilm biomass, detecting live and dead cells, as well as the exopolymeric matrix (Costa et al., 2018). The major problem with the crystal violet assay is its variability and, thus, variations in the data due to the user performing the assay (Lourenco et al., 2012) and the high detection limit. Better reproducibility of data can be achieved with live cell methods. Since transition of Listeria cells to a viable-but-non-culturable state is also possible under stress conditions (Highmore et al., 2018), the disadvantage of the CFU counting assay was that it only determined culturable live cells. In contrast, all live cells were determined using the resazurin fluorescence assay, which is based on the detection of metabolically active cells (Van den Driessche et al., 2014). This method was less sensitive, requiring at least 106 CFU/mL to detect a change in fluorescence.
The NanoLuc luciferase biofilm assay is a viable complement or addition to existing methods. Limitations of this method include the requirement of genetic modification of the bacteria and the high cost, even though the cost is lower than that for the molecular biology methods (e.g., qPCR). On the other hand, the Nluc bioluminescence assay has several important advantages over other methods, including direct detection, absolute cell quantification, broad dynamic range (1.0 × 104 CFU/mL – 5.0 × 107 CFU/mL), low time requirement, and high sensitivity (1.0 × 104 CFU/mL).
The protocol can be adapted to other bacteria by appropriate modification of the genetic construct (plasmid or promoter). Due to its high sensitivity, it is particularly useful for studies on early steps of biofilm development and for the bacteria that do not form thick biofilms. It can be easily applied to the screening of substances for antibiofilm activity.
Materials and Reagents
White flat-bottomed 96-well plates (Corning, catalog number: 3990)
15 mL centrifuge tube, PP, screw cap (Brand, catalog number: 114818)
1.5 mL microcentrifuge tubes (Eppendorf Safe-Lock, catalog number: EP0030123611)
Filtermax filter top (TPP, catalog number: 99505)
Presterile 50 mL disposable reservoirs (Biotix, catalog number: SR-0050-1SC)
Listeria innocua ŽM39 transformed with pMSP::Nluc (Jožef Stefan Institute collection) (Figure 1)
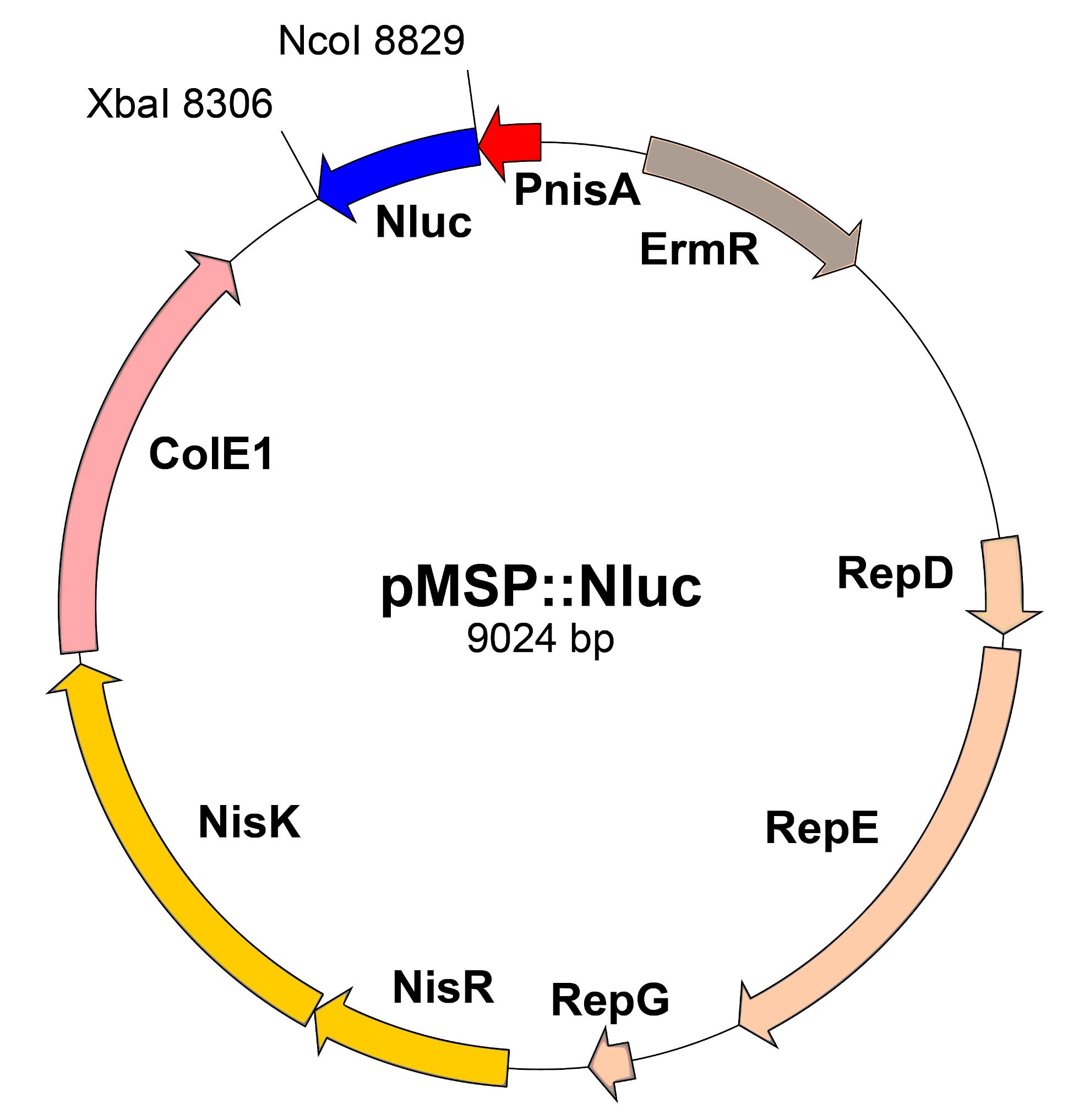
Figure 1. Scheme of pMSP::Nluc plasmid. PnisA: nisin promoter. ErmR: erythromycin resistance marker. RepD-G: replication genes. NisRK: nisin response genes. ColE1: E. coli replication gene. Nluc: Nanoluc.Tryptic soy broth (Merck, catalog number: 1.05459.0500)
Agar (Formedium, catalog number: AGA03)
Erythromycin (Merck, Sigma-Aldrich, catalog number: E5380)
Nisin (Merck, Sigma-Aldrich, catalog number: 72535)
Nano-Glo Luciferase Assay System (Promega, catalog number: N1120)
NaCl
KCl
Na2HPO4·2H2O
KH2PO4
Phosphate-buffered saline (PBS) (see Recipes)
TSA (see Recipes)
Equipment
Incubator (Binder)
Shaker/Incubator (Braun Certomat H)
Luminescence plate reader (Tecan, model: Infinite M-1000)
Pipette set (Eppendorf, catalog number: Z683892)
Multichannel pipette (Eppendorf, catalog number: 2231300048; model: Research Plus)
Biosafety cabinet (Labcaire)
Deep freezer (Sanyo, model: MDF-U53V)
Spectrophotometer (Perkin Elmer, catalog number: L7110187; model: Lambda Bio+)
Centrifuge (Hettich, catalog number: 1706; model: Rotina 380R)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Berlec, A., Janež, N., Sterniša, M., Klančnik, A. and Sabotič, J. (2022). Listeria innocua Biofilm Assay Using NanoLuc Luciferase. Bio-protocol 12(3): e4308. DOI: 10.21769/BioProtoc.4308.
分类
微生物学 > 微生物生物膜
微生物学 > 病原体检测 > 生物发光
生物科学 > 微生物学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link













