- EN - English
- CN - 中文
Measuring Oligonucleotide Hydrolysis in Cellular Lysates via Viscosity Measurements
通过粘度测量细胞裂解液中寡核苷酸的水解
发布: 2022年01月20日第12卷第2期 DOI: 10.21769/BioProtoc.4304 浏览次数: 2568
评审: Alba BlesaMiaowei Marwell MaoAnonymous reviewer(s)
Abstract
Cell lysis, a process that releases host oligonucleotides, is required in many biotechnological applications. However, intact oligonucleotides in crude cellular lysates increase the viscosity of lysates, which complicates downstream processes and routine laboratory workflows. To address this, nucleases that hydrolyze the intact oligonucleotides are commonly added, either as purified enzymes or co-expressed in genetically engineered bacterial strains. To measure oligonucleotide hydrolysis, common DNA quantification methods, such as qPCR or fluorescence-based, require expensive reagents and equipment, and cannot distinguish different-sized DNA fragments. Here, we outline a simple alternative method for measuring DNA/RNA hydrolysis in cellular lysates, by measuring their viscosity. This method only requires common laboratory supplies and a cell phone camera.
Background
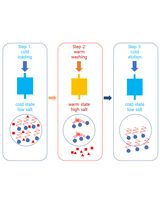
Many biotechnological applications, such as recombinant protein purification, require lysing the cells releasing host oligonucleotides in the process. However, intact oligonucleotides in crude cellular lysates rapidly increase the viscosity of biological samples, which can complicate sample handling and downstream processes (Prazeres et al., 1999), or may completely impede routine analytical methods, such as SDS-PAGE (Kurien and Scofield, 2012). While some cell lysis methods such as sonication lead to some DNA shearing and decreased viscosity, the addition of purified nucleases that hydrolyze intact oligonucleotides in lysates is a more commonplace solution in industrial settings (Boynton et al., 1999). Alternatively, we recently developed a method using an E. coli autolysis/autohydrolysis strain where, after cell growth and autoinducible protein expression, cell autolysis and DNA/RNA autohydrolysis can easily be triggered upon addition of a 0.1% detergent and a freeze-thaw cycle. This method leverages a chromosomally integrated genetic circuit that encodes the co-expression of a lysozyme and a nuclease (Menacho-Melgar et al., 2020).
In this context, a method to quantify oligonucleotide hydrolysis may be desirable. DNA quantification methods, such as fluorescence-based and qPCR, may be used for this purpose but require expensive equipment and reagents and do not differentiate based on DNA fragment size (Nakayama et al., 2016). A simpler alternative is to directly measure the viscosity of the lysates. While the viscosity of lysates is dependent on various factors, intact oligonucleotides are the major contributor. Still, viscosity can only be used to measure oligonucleotide hydrolysis within equivalent samples and not to make comparisons across different strains or cell cultures of different cell densities. Importantly, this method will not work in lysates obtained using sonication, as this shears the DNA. Here, we provide a detailed protocol for measuring DNA hydrolysis using viscosity measurements, only requiring the use of common laboratory supplies and a cell phone camera.
Materials and Reagents
5 mL serological pipets (Genesee Scientific, catalog number: 12-104)
Rattler glass plating beads (Genesee Scientific, catalog number: 11-255A)
Parafilm (VWR, catalog number: P7793-1EA)
General purpose laboratory tape (VWR, catalog number: 89097-938)
50 mL tubes (Genesee Scientific, catalog number: 28-108)
1.7 mL microcentrifuge tubes (Genesee Scientific, catalog number: 24-281)
Cell lysate samples to be tested (e.g., from DLF_R004) (Menacho-Melgar et al., 2020)
We recommend using a minimum of 2.5 mL of lysate, coming from at least 20 mL of cell culture, of at least OD600 = 10)
Tap water
Tris base (GoldBio, catalog number: T-400-500)
Triton X-100 (Sigma Aldrich, catalog number: 93443)
Protease inhibitors tablets, EDTA-free (Thermo Fisher, catalog number: A32965)
Lysis buffer (see Recipes)
Equipment
Cell phone with a camera
Tube cutter (Ridgid, model: 104)
Benchtop centrifuge (Thermo Scientific, model: Legend XTR)
Heat block (Bioer, model: CHB-201)
-80°C freezer (Thermo Scientific, model: TSX700)
Software
Any video media player such as “Movies & TV” (Microsoft)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Menacho-Melgar, R. and Lynch, M. D. (2022). Measuring Oligonucleotide Hydrolysis in Cellular Lysates via Viscosity Measurements . Bio-protocol 12(2): e4304. DOI: 10.21769/BioProtoc.4304.
分类
生物工程 > 合成生物学
微生物学 > 异源表达系统 > 大肠杆菌
生物化学 > 蛋白质 > 分离和纯化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link