- EN - English
- CN - 中文
Simple Scalable Protein Expression and Extraction Using Two-stage Autoinducible Cell Autolysis and DNA/RNA Autohydrolysis in Escherichia coli
利用两阶段自诱导细胞自溶和DNA/RNA自水解在大肠杆菌中表达和提取蛋白
发布: 2022年01月20日第12卷第2期 DOI: 10.21769/BioProtoc.4297 浏览次数: 3546
评审: Anonymous reviewer(s)

相关实验方案

重组人线粒体RNA聚合酶(POLRMT)及启动因子TFAM和TFB2M的表达和纯化
An H. Hsieh [...] Tatiana V. Mishanina
2023年12月05日 2498 阅读
Abstract
Recombinant protein expression is extensively used in biological research. Despite this, current protein expression and extraction methods are not readily scalable or amenable for high-throughput applications. Optimization of protein expression conditions using traditional methods, reliant on growth-associated induction, is non-trivial. Similarly, protein extraction methods are predominantly restricted to chemical methods, and mechanical methods reliant on expensive specialized equipment more tuned for large-scale applications. In this article, we outline detailed protocols for the use of an engineered autolysis/autohydrolysis E. coli strain, in two-stage fermentations in shake-flasks. This two-stage fermentation protocol does not require optimization of expression conditions and results in high protein titers. Cell lysis in an engineered strain is tightly controlled and only triggered post-culture by addition of a 0.1% detergent solution. Upon cell lysis, a nuclease digests contaminating host oligonucleotides, which facilitates sample handling. This method has been validated for use at different scales, from microtiter plates to instrumented bioreactors.
Graphic abstract:
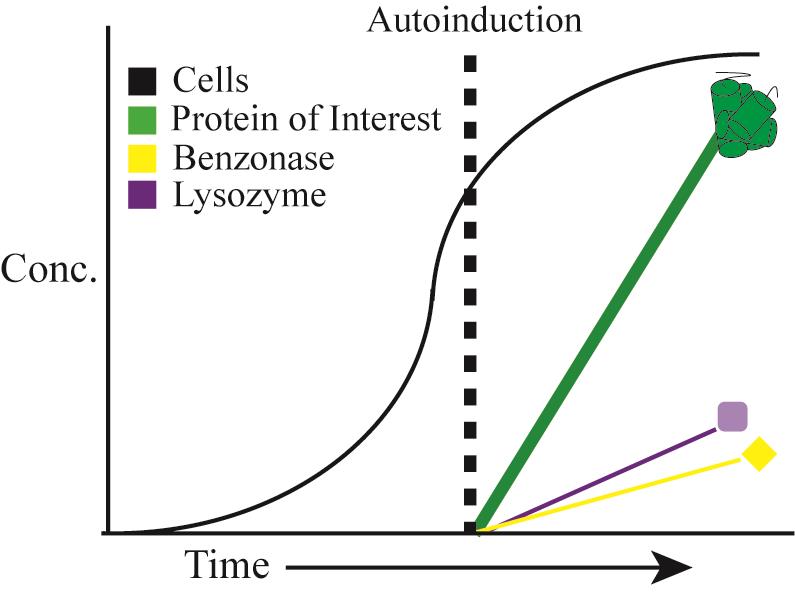
Two-stage protein expression, cell autolysis and DNA/RNA autohydrolysis. Reprinted with permission from Menacho-Melgar et al. (2020a). Copyright 2020 John Wiley and Sons.
Background
Despite heterologous protein expression being widely used in biotechnological research, optimization of protein expression protocols can be a very lengthy and costly process (Bill, 2014; Rosano and Ceccarelli, 2014). Once determined, optimal expression conditions are not readily translated to other fermentation scales or proteins of interest, thereby limiting method transfer to industrial scales, as well as increasing process development timelines (Mühlmann et al. 2017). After protein expression, many applications often require protein purification that calls for using expensive specialized equipment. Specifically, cell lysis for protein extraction is still mostly performed using mechanical methods, such as sonication or homogenization, which may not yield consistent results and limit high-throughput applications (Foster, 1992; Cai et al., 2008). Alternative methods, such as chemical methods orengineering “autolysis” microbial strains that lyse after cell harvesting, exist but are poorly controlled and do not account for oligonucleotides removal to reduce sample viscosity, which can complicate sample handling (Chien and Lee, 2006; Cai et al., 2008).
To address these, we recently described a two-stage protein expression process (Menacho-Melgar et al., 2020b). In this method, we use engineered strains that do not produce organic acid byproducts (e.g., acetate), known to negatively affect cell growth and protein expression (Jian et al., 2010), in combination with plasmids, where the expression of the protein of interest is under the tight control of a low phosphate inducible promoter (refer to Figure 1 for an example annotated plasmid sequence) (Moreb et al., 2020). In two-stage fermentations, protein expression is triggered upon entry into the stationary phase, when phosphate concentration in the media becomes depleted, which (i) stops cell growth (stage 1) and (ii) induces protein expression (stage 2) (Figure 2A). Two-stage fermentations decouple cell growth from protein expression, which results in high protein titers and enables the use of already optimized standard protocols across different scales and proteins of interest (Burg et al., 2016; Decker et al., 2020). In other words, two-stage protein expression does not require optimizing induction conditions, as opposed to methods that induce protein expression during growth. Additionally, this approach has enabled higher protein titers than growth-associated protein expression.
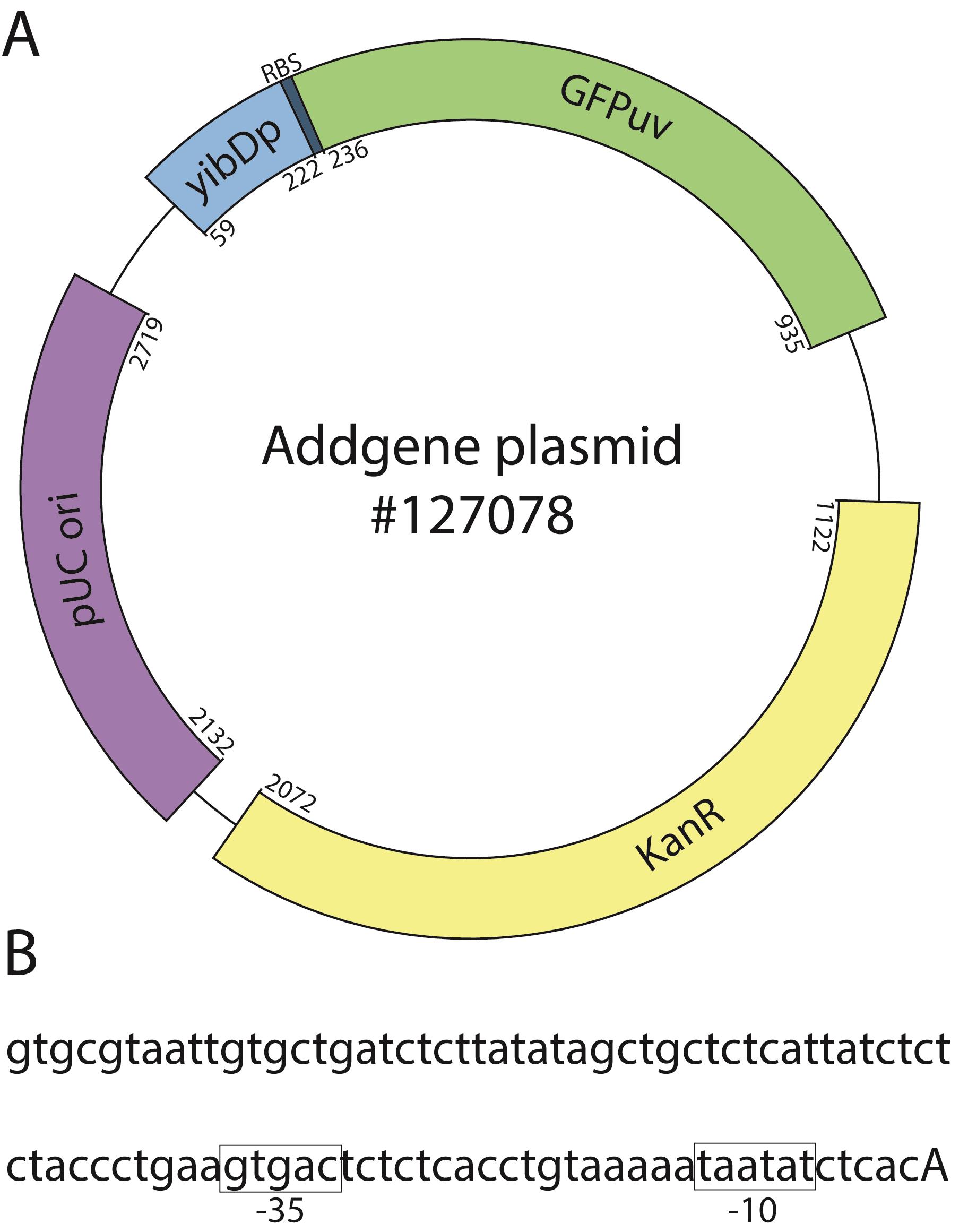
Figure 1. Example two-stage expression plasmid. A. Expression of the protein of interest (GFPuv, green) is driven by a low-phosphate inducible promoter (yibDp, blue). The plasmid has the high copy pUC origin (pBR322 derivative, purple) and a kanamycin resistance marker (yellow). B. Partial yibD promoter sequence showing the -10 and -35 boxes (highlighted in boxes) and the start of transcription (capitalized). Start and end bases for each feature are annotated. For a complete plasmid sequence, see https://www.addgene.org/browse/sequence/271197/ under the ‘Sequence’ tab.
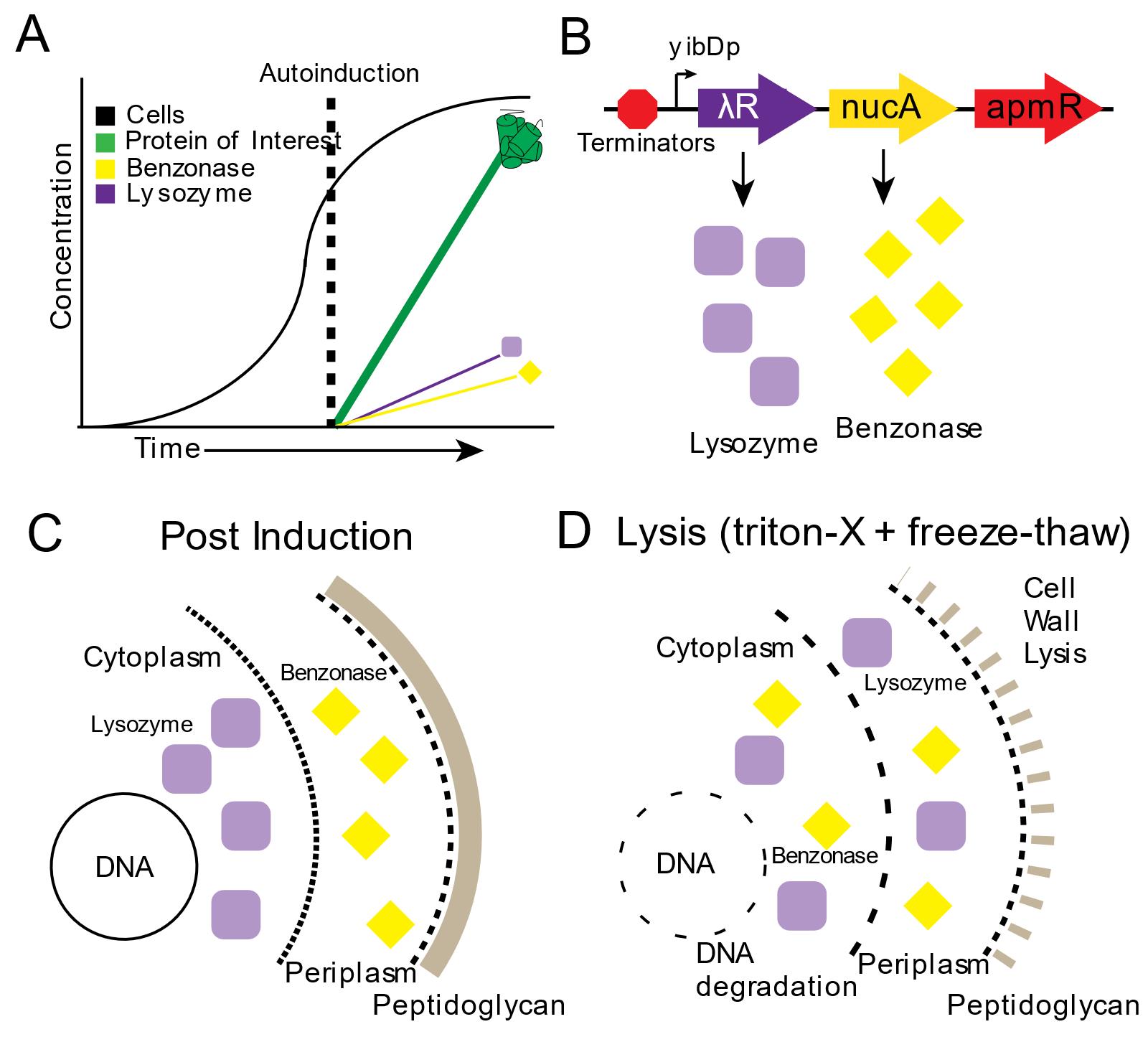
Figure 2. Autolysis and Autohydrolysis in two-stage protein expression. A. In stage 1 “growth”, cells are grown in AB-2 media. In stage 2, “production” gets triggered upon phosphate depletion in the media, when cells enter the stationary phase, activating expression of the protein of interest. B. Expression of the autolysis/autohydrolysis machinery, that is chromosomally integrated and consists of benzonase and lysozyme, also occurs at this stage. C. After induction, lysozyme remains inside the cytoplasm and benzonase is located in the periplasmic space. D. Only upon addition of detergent (Triton-X) and a freeze-thaw cycle, the cells lyse and the endonuclease is able to digest the host’s DNA. Reprinted with permission from Menacho-Melgar et al. (2020a). Copyright 2020 John Wiley and Sons.
Additionally, we built upon two-stage protein expression and engineered an autolysis/autohydrolysis strain, which expresses a lysozyme (from the lambda R gene) in the cytoplasm and a nuclease (from S. marcescens nucA gene, commercially known as BenzonaseTM) in the periplasm, during the protein production stage (Figure 2A-2C) (Menacho-Melgar et al., 2020a). These chromosomally integrated enzymes do not negatively affect the titer of the protein of interest (Menacho-Melgar et al., 2020a). Since this system is tightly controlled, no cell lysis will occur during the growth phase or after harvesting, as opposed to other BL21-based autolysis strains (Menacho-Melgar et al., 2020a). Only upon resuspension in a lysis buffer containing 0.1% detergent and a freeze-thaw cycle, over 90% of the protein is released (compared to a 20-minute sonication protocol, where further sonication does not lead to more protein release) and all of the oligonucleotides in the cell lysate are removed (Figure 2D) (Menacho-Melgar et al., 2020a). Importantly, this method enables the simultaneous lysis of several samples, which can be essential in high-throughput applications (Menacho-Melgar et al., 2020a). Although this method has been validated across different fermentation scales (Menacho-Melgar et al., 2020a), fermentations in microtiter plates and bioreactors require specialized equipment and accessories not readily accessible, whereas fermentations in shake-flasks are more widely and routinely used. Thus, in this protocol, we have described a detailed protocol for performing two-stage fermentations only in shake flasks, as well as protein extraction using our autolysis/autohydrolysis strain and a plasmid expressing GFPuv (Addgene, #127078), as an example (Menacho-Melgar et al., 2020a).
Materials and Reagents
10, 200, and 1,000 μL pipette tips (Genesee Scientific, catalog numbers: 23-121RLC, 23-150RLC and 23-165RLC)
1.7 mL microcentrifuge tubes (Genesee Scientific, catalog number: 24-281)
15 mL culture tubes (Genesee Scientific, catalog number: 21-130)
50 mL conical tubes (Genesee Scientific, catalog number: 28-108)
2 mL cryovials (VWR, catalog number: 10018-754)
1 L media bottles (VWR, catalog number: 10754-820)
10 mL and 25 mL serological pipets (Genesee Scientific, catalog numbers: 12-104 and 12-106)
250 vented baffled shake flask (VWR, catalog number: 89095-270)
Tryptone (Biobasic, catalog number: TG217(G211))
Yeast extract (Biobasic, catalog number: G0961)
Sodium chloride (Biobasic, catalog number: DB0483)
Ammonium sulfate anhydrous (VWR, catalog number: M105)
Glucose (VWR, catalog number: 89405-376)
Bis-Tris (GoldBio, catalog number: B-020-500)
Casamino acids (Biobasic, catalog number: CB3060)
12 M Hydrochloric acid (Biobasic, catalog number: HC6025)
Tris base (GoldBio, catalog number: T-400-500)
Triton X-100 (Sigma Aldrich, catalog number: 93443)
Protease inhibitors tablets EDTA-free (A32965)
Strain DLF_R004 (F-, λ-, Δ(araD-araB)567, lacZ4787(del)::rrnB-3), rph-1, Δ(rhaD-rhaB)568, hsdR51, ΔackA-pta, ΔpoxB, ΔpflB, ΔldhA, ΔadhE, ΔicIR, ΔarcA, ΔompT::yibDp- λR-nucA-apmR) transformed with a plasmid expressing your protein of interest (e.g., GFPuv), under a low phosphate inducible promoter (e.g., yibD) (e.g., Addgene plasmid #127078)
Low salt LB (see Recipes)
AB-2 (see Recipes)
Lysis buffer (see Recipes)
Equipment
Pipette controller (Eppendorf, model: 4430 000 018)
Incubator shaker, orbit set to 50 mm (Kuhner, model: ISF4-X)
Benchtop centrifuge (Thermo Scientific, model: Legend XTR)
-80°C freezer (Thermo Scientific, model: TSX700)
Microcentrifuge (Eppendorf, model: 5415R)
Heat block (Bioer, model: CHB-201)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Menacho-Melgar, R. and Lynch, M. D. (2022). Simple Scalable Protein Expression and Extraction Using Two-stage Autoinducible Cell Autolysis and DNA/RNA Autohydrolysis in Escherichia coli . Bio-protocol 12(2): e4297. DOI: 10.21769/BioProtoc.4297.
分类
微生物学 > 异源表达系统 > 大肠杆菌
分子生物学 > 蛋白质 > 表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link