- EN - English
- CN - 中文
High-throughput Growth Measurements of Yeast Exposed to Visible Light
酵母暴露于可见光下的高通量生长测量
发布: 2022年01月20日第12卷第2期 DOI: 10.21769/BioProtoc.4292 浏览次数: 3175
评审: Gal HaimovichIndranil MalikAnonymous reviewer(s)
Abstract
Light is a double-edged sword: it is essential for life on the planet but also causes cellular damage and death. Consequently, organisms have evolved systems not only for harvesting and converting light energy into chemical energy but also for countering its toxic effects. Despite the omnipresence and importance of such light-dependent effects, there are very few unbiased genetic screens, if any, investigating the mechanistic consequences that visible light has on cells. Baker’s yeast, Saccharomyces cerevisiae, is one of the best annotated organisms thanks to several easily available mutant collections and its amenability to high-throughput genetic screening. However, until recently this yeast was thought to lack receptors for visible light, therefore its response to visible light was poorly understood. Nevertheless, a couple of years ago it was discovered that yeast senses light via a novel and unconventional pathway involving a peroxisomal oxidase, hydrogen peroxide, and a particular type of antioxidant protein, called peroxiredoxin. Here, we describe in detail a protocol for scoring yeast genes involved in the resistance to visible light (400-700 nm) on a genome-wide scale. Because cells in dense cultures shield each other from light exposure, resulting in apparent light resistance, our method involves adaptations to reduce inoculum size under conditions amenable to high-throughput screens, to properly be able to identify light-sensitive mutants. We also describe how to measure growth in the presence of light, including two follow-up validation tests. In this way, this method makes it possible to score light-sensitivity on a genome-wide scale with high confidence.
Graphic abstract:
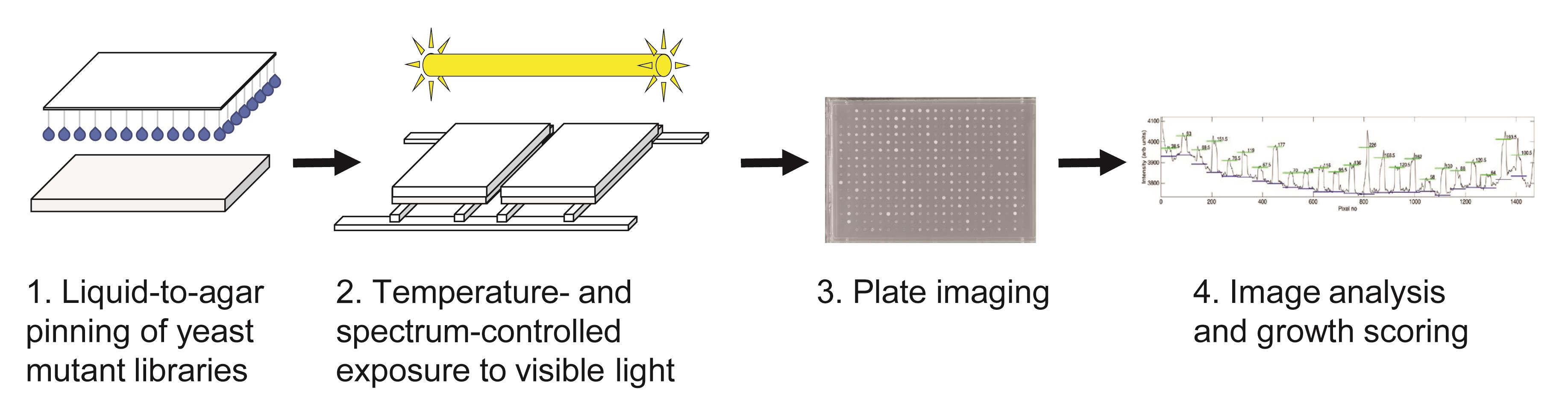
Overview of strategy for high-throughput determination of yeast growth upon visible light stress.
Background
Light is, on the one hand, essential for life on the planet as a source of energy but, on the other hand, it can also be involved in regulatory processes, as well as cause substantial cellular damage and demise. Studies in later years have suggested that cells from the three domains of life, including most human cells, keep stimulus-independent rhythms of activity to adapt to the prevailing recurring alterations of sunlight and darkness (i.e., circadian rhythms). A key stimulus entraining circadian rhythms is visible light. Characterized clock mechanisms, including those in filamentous fungi, involve cryptochrome, rhodopsin, and melanopsin photoreceptors (Yu and Fischer, 2019; Patke et al., 2020). However, circadian rhythms persist in red blood cells in the absence of canonical clock components (O'Neill and Reddy, 2011; O'Neill et al., 2011), indicating that alternative clock mechanisms must exist. In red blood cells, the unusual anti-oxidants and signaling regulators peroxiredoxins were proposed to constitute conserved and transcription-independent regulators and markers of circadian rhythms (Edgar et al., 2012). On a similar note, baker’s yeast has long been thought to lack light receptors (Idnurm et al., 2010). Recently, however, it was discovered that yeast can sense light via a peroxisomal oxidase, hydrogen peroxide, and the peroxiredoxin Tsa1 (Bodvard et al., 2017). Thus, yeast is a model for understanding responses to light in cells and organisms lacking dedicated light receptors. In the yeast Saccharomyces cerevisiae, high-throughput genetic screening by ordered arrays of mutant libraries has become a central tool in understanding gene function on a genome-wide scale (Giaever et al., 2002).
Several filamentous fungi are able to sense light of different wavelengths through photoreceptors homologous to those in mammals and plants. In fungi, light controls circadian clocks and morphological responses, as well as growth/stress response decisions. Investigations of their responses to light have mainly focused on identifying photoreceptors, circadian clock components, and regulators of morphological responses. In these organisms, large-scale investigations of light responses have so far been limited to transcriptomics (Lewis et al., 2002; Dong et al., 2008; Chen et al., 2009; Wu et al., 2014), forward genetics screens using light-dependent reporters (Yu et al., 2016), and reverse genetic screens of limited gene sets (e.g., genes encoding transcription factors) affecting circadian clock reporters (Munoz-Guzman et al., 2021). Here, we outline a protocol for assessing baker’s yeast functions relevant in responses to visible light (400-700 nm) on a genome-wide scale, by scoring light-dependent growth of mutants. Potentially, by using suitable reporters (Kainth et al., 2009; D'Orazio et al., 2019) and with proper modifications to the setup, this protocol could be used to study other types of responses to light or, depending on the availability of genome-wide mutant collections, also understand light-resistance in other microorganisms.
The procedure we describe here, to score genes involved in the resistance of yeast to visible light, starts at a genome-wide scale, and subsequently narrows down to smaller scale validation tests for selected light-sensitive strains. In our case, the haploid BY4741 deletion collection was examined, in an arrayed format used in the Boone lab for synthetic genetic array analysis (Tong et al., 2001), in which all plate edges are lined by a row of wild-type-like (his3∆) control strains. We describe a detailed protocol for achieving the initial genome-wide screen, despite a strong dependence of yeast light-dependent growth on cell density that complicated analyses. Furthermore, to be able to reliably score light-sensitivity, we next describe appropriate methods for image analysis and growth scoring in detail. Following this, we describe a medium-throughput assay suitable for retesting a hundred or so strains, which should preferably be used on multiple deletion mutant collections, such as those containing the haploid (BY4741) and homozygous diploid (BY4743) gene knockouts. Finally, we outline a serial dilution drop test assay suitable for a smaller subset of genes, which in our case was used to retest gene deletions from the homozygous diploid BY4743 collection.
Materials and Reagents
96 well plates: TPP® Tissue culture plate 96 well, flat-bottom (Merck, catalog number: Z707902)
Experimental agar plates: PlusPlates (Singer Instruments, catalog number: PLU-003)
Pinning pads: RePads 96 long pin (Singer Instruments, catalog number: REP-001)
14 mL Falcon® tubes round-bottom with snap-cap (VWR, catalog number: 734-0985)
Yeast haploid deletion mutant collection (BY4741 (Mat a) transOMIC technologies, catalog number: TKY3502)
Yeast homozygous diploid deletion mutant collection (BY4743, transOMIC technologies, catalog number: TKY3500)
Glucose (Sigma-Aldrich, catalog number: G8270)
Peptone (VWR, catalog number: ICNA0210480805)
Yeast extract (VWR, catalog number: J850)
Yeast Nitrogen Base without Amino Acids and without Ammonium Sulphate (Formedium, catalog number: CYN0510)
Agar (VWR, catalog number: J637)
Ammonium sulfate (Sigma-Aldrich, catalog number: A4418)
Complete Supplement Mixture with all amino acids (Formedium, catalog number: DCS0019)
Succinic acid (Sigma Aldrich, catalog number: 398055)
NaOH (Sigma Aldrich, catalog number: S5881)
40% (w/w) glucose (see Recipes)
YPD liquid medium (see Recipes)
10× stock of amino acid mixture (see Recipes)
10× stock of succinate buffered Yeast Nitrogen Base (YNB) (see Recipes)
10× stock of amino acid mixture (see Recipes)
Ultrapure (Milli-Q) water (see Recipes)
Agar mixture (see Recipes)
Equipment
For shaking plates (Vibrax VXR basic, IKA, Germany)
Robot for pinning (RoToR HDA; Singer Instruments, UK)
Open industrial light tube fitting for 3 × 58 W light tubes (Mir, Elektroskandia, product number E72 106 41)
UV radiation-filtered fluorescent light tubes (Osram L 58 W/940)
Sample holders: for each plate two ribs holding the plate above ground to allow for air circulation and specific distance from lamp
Spectrometer (QE65000-FL, Oceans Optics)
Imaging of plates (Digital camera D5000 Nikon and Epson Perfection V700)
Software
MATLAB® (for high- and medium-throughput assays)
QuantityOne (Bio-Rad, for serial dilution drop test)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Logg, K., Andersson, M., Blomberg, A. and Molin, M. (2022). High-throughput Growth Measurements of Yeast Exposed to Visible Light . Bio-protocol 12(2): e4292. DOI: 10.21769/BioProtoc.4292.
分类
微生物学 > 微生物生理学 > 胁迫反应
微生物学 > 微生物信号传导 > 感知受体
细胞生物学 > 基于细胞的分析方法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link