- EN - English
- CN - 中文
Quantification of Bacterial Loads in Caenorhabditis elegans
秀丽隐杆线虫细菌负荷的定量研究
发布: 2022年01月20日第12卷第2期 DOI: 10.21769/BioProtoc.4291 浏览次数: 4229
评审: Ambre SalaAnthony OrtizAnonymous reviewer(s)
Abstract
Caenorhabditis elegans is a ubiquitous free-living nematode that feeds on bacteria. The organism was introduced into a laboratory setting in the 1970s and has since gained popularity as a model to study host-bacteria interactions. One advantage of using C. elegans is that its intestine can be colonized by the bacteria on which it feeds. Quantifying the bacterial load within C. elegans is an important and easily obtainable metric when investigating host-bacteria interactions. Although quantification of bacteria harbored in C. elegans via whole-worm lysis is not a novel assay, there is great variation between existing methods. To lyse C. elegans, many protocols rely on the use of a hand-held homogenizer, which could introduce systematic error and subsequent variation between researchers performing the same experiment. Here, we describe a method of lysing the intestines of C. elegans to quantify the bacterial load within the intestine. Our method has been optimized for removing exogenous bacteria while maintaining worm paralysis, to ensure no bactericidal agents are swallowed, which could kill bacteria within the intestine and affect results. We utilize and compare the efficiency of two different homogenization tools: a battery-powered hand-held homogenizer, and a benchtop electric homogenizer, where the latter minimizes variability. Thus, our protocol has been optimized to reduce systematic error and decrease the potential for variability among experimenters.
Graphic abstract:
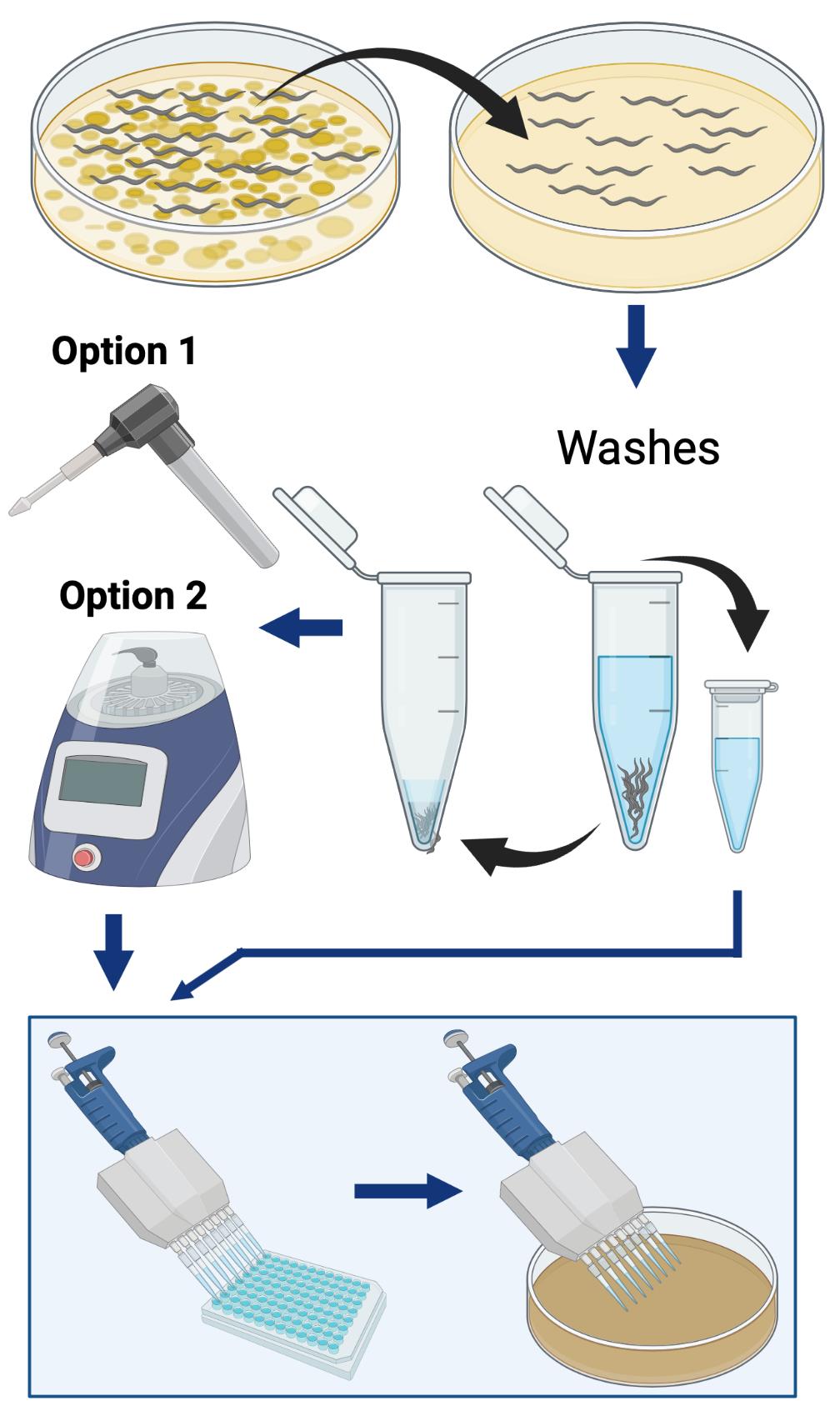
Simplified overview of the procedure used to quantify the bacterial load within C. elegans. The two different methods are herein described for worm lysis: “Option 1” is a hand-held homogenizer, and “Option 2” is a benchtop homogenizer.
Background
The nematode Caenorhabditis elegans is a bacterivore that is widely used as a model to study host-bacteria interactions, due to its simple morphology, high homology of its genome to the human genome, and the amenability of its microbiome (Zhang et al., 2017). These nematodes use their pharynx to intake bacteria from their surrounding environment (Avery and You, 2018). In a laboratory setting, germ-free larvae are obtained by isolating embryos from gravid adults. Once hatched, larvae (L1s) will begin to feed on bacteria (Brenner, 1973). Hence, it is easy to colonize the C. elegans gut with bacterial strain(s) of interest. Studies involving host-bacteria interactions often call for the enumeration of the bacterial load within the intestine. One of the most common and reliable bacterial quantification techniques is plating for colony forming units (CFUs). Quantification of bacteria within C. elegans is achieved via the physical disruption of worm bodies to release intestinal content, and plating the resulting lysate on solid medium to support microbial growth and subsequent enumeration of CFUs. However, there is variation between existing protocols for the aforementioned assay. For example, prior to the homogenization process, worms are washed several times in a microcentrifuge tube to eliminate external bacteria from the sample. During these washes, worms must be allowed to settle to the bottom of the tube before the supernatant is aspirated, and the researcher must be aware of the location of the worms within the tube, to ensure worms are not aspirated out with the supernatant—because worms tend to stick to pipette tips, this is a critical precautionary measure, which is not emphasized in many existing protocols. Many protocols also call for gentamicin or bleach washes, to eliminate bacteria attached to the cuticle of the worm prior to homogenization. However, if swallowed, bactericidal agents could affect the viability of intestinal bacteria, skewing results. To circumvent these risks, we add levamisole, an alkaline phosphatase inhibitor that reversibly paralyzes C. elegans, to our gentamicin and non-gentamicin washing solutions. This step stops pharyngeal pumping and decreases the risk of the germicidal agent making its way to the intestine. Other techniques used to prevent ingestion of bactericidal agents include lowering the temperature of the samples, and adding reagents that retard motility and minimize pharyngeal pumping (Vega and Gore, 2017; Ortiz et al., 2021). Temperature-mediated inhibition of motility is an attractive approach that can be incorporated into our protocol, for applications that may want to avoid using levamisole. Manual homogenization of worms introduces the most variability. For example, moving the pestle rapidly in the sample tube could cause bubbling and subsequent spillage of the sample. The time and force exerted on the pestle could also affect reproducibility between experimenters. We describe detailed steps in our protocol, such as the position of the pestle and the movement of the hand-held homogenizer, to mitigate the aforementioned risks. Other more hands-off methods of disrupting worm bodies include the automated higher-throughput worm lysis that likely limits the risk of human-induced variation (Vega and Gore, 2017). As such, we describe a benchtop homogenizer-based automated method that diminishes the risk of human error and variation. A flow-chart of the described method is illustrated in Figure 1. Three researchers in our lab performed the protocol described herein on the same day, using worms taken from the same sample. Quantification of the bacterial load within C. elegans varied between researchers using a hand-held homogenizer (Figure 2A), whereas results were nearly identical when a benchtop homogenizer was used (Figure 2B). Hence, option 2 should be the method of choice, as it provides the most consistent results. It is worth noting that this protocol can be modified to permit quantification of other microbes harbored within C. elegans, or any experiment that requires worm lysis to be consistent between samples.
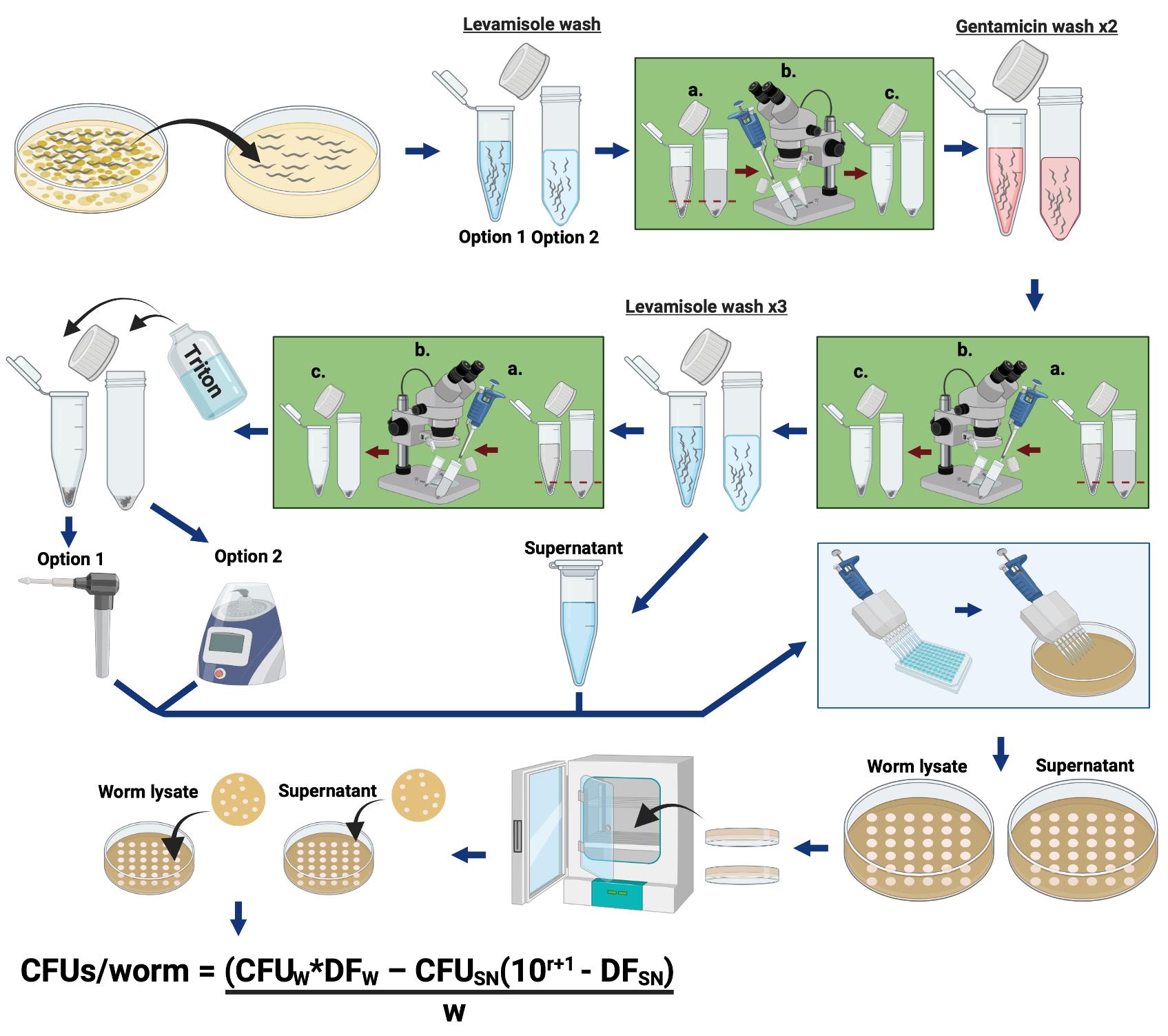
Figure 1. Schematic illustrating the protocol “Quantification of Bacterial Loads in C. elegans”. Detailed methods can be found in “Procedure”. Explanation of CFU calculations can be found in “Data Analysis”.
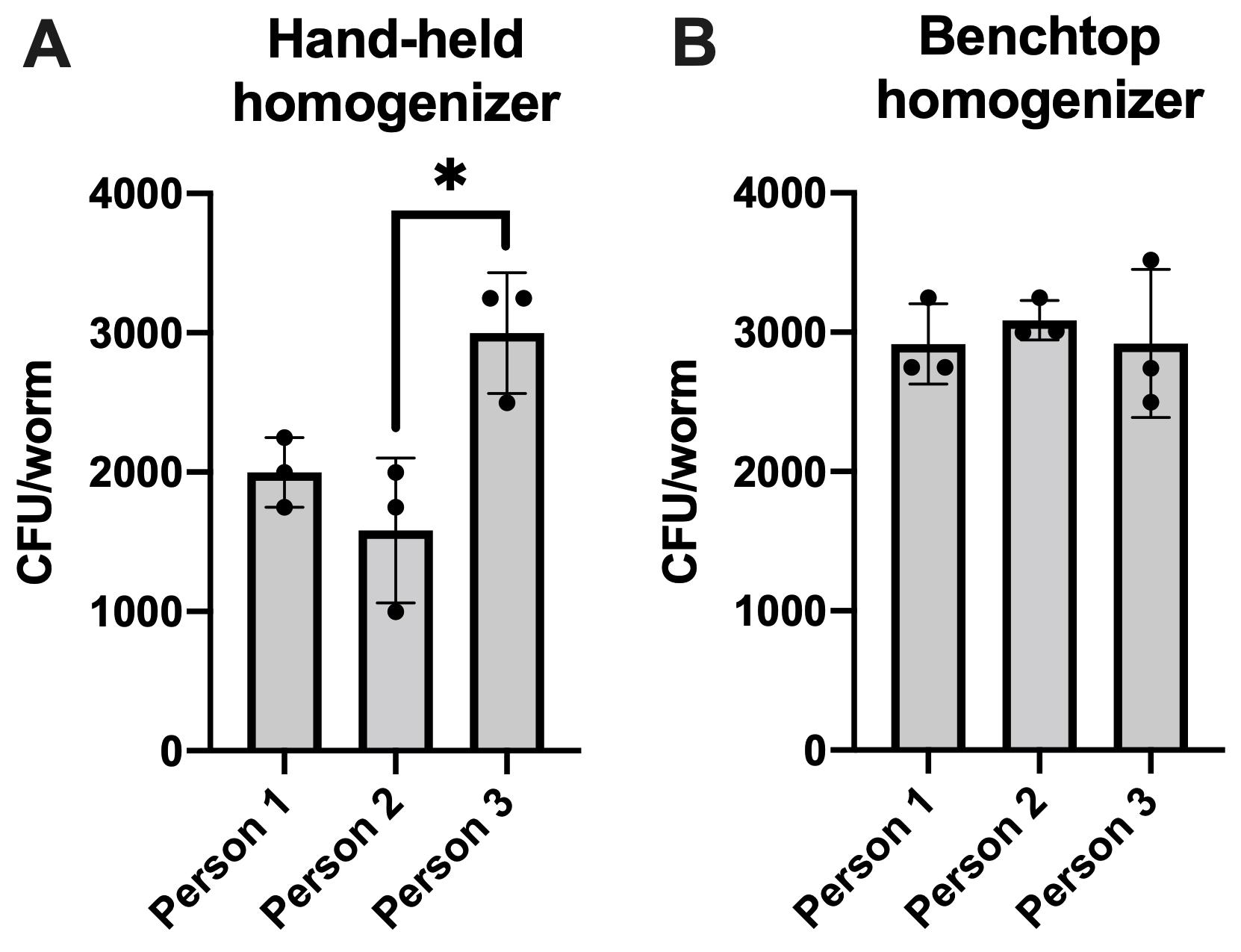
Figure 2. Quantification of bacteria within C. elegans. Reproducibility between three experimenters conducting the protocol described herein using (A) a hand-held homogenizer, and (B) a benchtop homogenizer to lyse C. elegans. Data are represented as the average bacterial load per C. elegans intestine. Each bar represents one independent experiment with three internal replicates. Error bars represent standard error of the mean (SEM). Significance was calculated using one-way analysis of variance (ANOVA) followed by multiple comparison Tukey’s post-hoc test (*P < 0.05).
Materials and Reagents
Note: Unless otherwise specified, all reagents are stored at room temperature.
Erlenmeyer flasks (2,000 mL and 1,000 mL)
6 cm Petri dishes (Genesee Scientific, catalog number: 32-105G)
10 cm Petri dishes (Genesee Scientific, catalog number: 32-107G)
Pestle and tube (Bel-Art F650000006, catalog number: 03-421-221), if following “Option 1”; or empty 2 mL non-skirted screw cap micro tubes with conical bottom (Thermo Scientific 3468, catalog number: 21-403-200), if following “Option 2” in Step C of “Procedure”
Bead Bug 1.5 mm zirconium beads (Benchmark Scientific, catalog number: D1032-15)
Pipette tips (P1000, P200, P20, or P10)
Magnetic stir bar
Autoclave tape
500 mL Olympus vacuum filter flasks (Genesee Scientific, catalog number: 25-227)
Lennox Luria broth powder (Genesee Scientific, catalog number: 11-125)
Agar (Fisher Scientific, catalog number: BP1423)
Double distilled water (ddH2O)
NaCl (Fisher Scientific, catalog number: S671-500)
Trypticase-peptone (Gibco, catalog number: 211921)
Cholesterol (MP Biomedicals, catalog number: ICN10138201)
Ethanol, 200-proof (Decon Laboratories, catalog number: 04-355-223)
CaCl2·2H2O (Fisher Scientific, catalog number: C79-500)
MgSO4·7H2O (Fisher Scientific, catalog number: A14491)
KH2PO4 (Fisher Chemical, catalog number: P285-3)
NA2HPO4·7H2O (Fisher Scientific, catalog number: S373-500)
Levamisole hydrochloride (MP biomedicals, catalog number: 155228)
Triton X-100 (Promega, catalog number: H5142)
Phosphate-buffered saline (PBS), 10× (Fisher Scientific, catalog number: J62036)
Gentamicin sulfate solution (Cassion Labs, catalog number: ABL03)
LB agar plates (see Recipes)
Phosphate Buffered Saline (PBS), 1× (see Recipes)
M9 Minimal Medium (see Recipes)
Levamisole Stock Solution (see Recipes)
M9/Levamisole Solution (see Recipes)
Gentamicin Stock Solution (see Recipes)
Gentamicin Wash Solution (see Recipes)
Cholesterol Stock Solution (see Recipes)
1 M CaCl2 Stock Solution (see Recipes)
1 M MgSO4 Stock Solution (see Recipes)
1 M KH2PO4, pH 6.0 Stock Solution (see Recipes)
NGM Plates (see Recipes)
1% Triton X-100 solution (see Recipes)
Equipment
Microtube Homogenizer System (Bel-Art, catalog number: F65000-0000)
BeadBug3 Microtube Homogenizer (Benchmark Scientific, catalog number: D1030)
Zeiss Stemi 305 stereo microscope (Zeiss, catalog number: 435063-9010-000)
Incubators for bacteria (37°C) and C. elegans (15-25°C)
Pipette (Eppendorf, models: P1000, P200 or P20, multichannel models P200, P10 or P20)
Platinum wire worm pick (made in-house as described in Wollenberg et al., 2013)
Ethanol candle (DWK Life Sciences, catalog number: 04-245-1)
Autoclave
Biological safety cabinet
Denver Instrument Summit SI-4002 Summit Series Toploading Balance (Denver instruments, catalog number: EW-11422-13)
pH meter (Thermo Electric Corporation)
Fisher Scientific Isotemp Stirrer (Fisher Scientific, catalog number: 11-100-49S)
Software
Microsoft Excel (http://office.microsoft.com/excel)
Graphpad Prism v8.4.3. (Graphpad Software, Inc., https://www.graphpad.com)
BioRender (www.biorender.com)
Procedure
文章信息
版权信息
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Walker, A. C., Bhargava, R., Vaziriyan-Sani, A. S., Brust, A. S. and Czyz, D. M. (2022). Quantification of Bacterial Loads in Caenorhabditis elegans. Bio-protocol 12(2): e4291. DOI: 10.21769/BioProtoc.4291.
分类
微生物学 > 微生物-宿主相互作用 > 线虫
微生物学 > 微生物-宿主相互作用 > 细菌
生物科学 > 微生物学 > 微生物群落 > 微生物组学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link