- EN - English
- CN - 中文
Measuring Real-time DNA/RNA Nuclease Activity through Fluorescence
通过荧光测量实时 DNA/RNA 核酸酶活性
发布: 2021年11月05日第11卷第21期 DOI: 10.21769/BioProtoc.4206 浏览次数: 3896
评审: Khyati Hitesh ShahMukesh MahajanAnonymous reviewer(s)
Abstract
DNA and RNA nucleases are wide-ranging enzymes, taking part in broad cellular processes from DNA repair to immune response control. Growing interest in the mechanisms and activities of newly discovered nucleases inspired us to share the detailed protocol of our nuclease assay (Sheppard et al., 2019). This easy and inexpensive method can provide data that enables understanding of the molecular mechanism for novel or tested nucleases, from substrate preference and cofactors involved to catalytic rate of reaction.
Keywords: Nuclease assay (核酸酶检测)Background
Nucleases are enzymes that act on DNA and RNA by cleaving the phosphodiester bonds between nucleotides. In addition to their crucial role in the DNA repair machinery, they are involved in cell signalling pathways pertinent to DNA damage and immune responses, among others (Sheppard et al., 2018). Their complex roles support several premature ageing-, immune-, and tumour-related processes. All of these can suffer from aberrations in the structural and/or catalytic functions of DNA and RNA nucleases (reviewed in Bartosova et al., 2014; Rigby et al., 2015). The growing interest in understanding the activities of numerous human DNA nucleases that remain contentious [e.g., Mre11 (Paull and Deshpande, 2014) and CTIP (Mozaffari et al., 2021)] and the presence of several uncharacterised proteins with predicted nuclease domains in mammalian genomes led us to design a real-time nuclease assay.
The activity and kinetics of nucleases, DNA polymerases, nickases, RNA:DNA nucleases, or single-strand DNA nucleases can be studied in an uncomplicated and cost-effective manner. The fluorescence signal changes resulting from decreasing amount of intercalated DNA dye can be quickly and safely measured by most plate readers. A wide range of oligonucleotides mimicking the DNA substrate of the tested enzyme can be examined in each experiment simultaneously. We designed and tested an oligomer library of 80mers with different characteristics and substrate potential. The oligonucleotides described allow for the determination of the enzymatic direction of nuclease activity. For example, 3′ or 5′ activity can be tested and compared with oligonucleotides containing biotin- blocked or free 3′ends or 5′ends, in addition to overhangs, gaps, or nicks. This method can also illustrate the importance of cofactors or cations through simple comparison between reactions supplemented or not with the chemical/cation. Using the same principle, a modified protein (phosphorylation, dephosphorylation) or mutated/truncated forms can be easily tested for their nuclease activity. Importantly, the assay is sensitive enough to detect the kinetics of repair enzymes when confronted with DNA mismatches or DNA methylation sites.
Materials and Reagents
Prepare all buffers and solutions using ultrapure, nuclease free-water and analytical grade reagents. Filter with 0.2 µm filter at least once and store at 4°C or -20°C. Always use nuclease-free tubes and cotton-filter tips.
Black bottom plates [black bottom plate 96 well, polypropylene, flat bottom (Chimney well)] are necessary for the fluorescence assay to reduce background and crosstalk, and to absorb light (Greiner Bio-One, catalog number: 655209)
Streptavidin (Pierce, catalog number: PIER21122)
Streptavidin (SA) has a great affinity to biotin triethyleneglycol (BITEG). Oligonucleotides with this modification on 3′, 5′, or both ends are protected from nuclease activity after adding 2 μl (0.02 mg/ml) streptavidin to the reaction mix. Prepare 1 mg/ml in ultrapure water.
Oligonucleotides
Order lyophilised unmodified HPLC-purified oligonucleotide substrates and biotin or BITEG modified HPLC-purified oligonucleotides (Integrated DNA Technologies). Dilute them in 1× Annealing Buffer (Sigma-Aldrich) for DNA substrates (100 μM stock) and siMAXTM dilution buffer (Eurofins; 30 mM HEPES, 100 mM KCl, 1 mM MgCl2, pH = 7.3) for all RNA substrates.
Note: Modified DNA or RNA bases can be ordered in the synthesised oligos. We have done so successfully in the past to measure the effect of base modifications on enzymatic functions.
Oligonucleotides from our library were designed and optimised against secondary structure formation using the ‘Predict a Secondary Structure Web Server’ (https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html).
The basic 80bp sequences are as follows:
Forward: GATGGTTTGTTGGTCTATTACTACTTGGAGCTTGTATGATTCGAAACCTTGGAGTACTTGCCTACTTGGAGTGAACTTAG
Reverse: CTAAGTTCACTCCAAGTAGGCAAGTACTCCAAGGTTTCGAATCATACAAGCTCCAAGTAGTAATAGACCAACAAACCATC
For designing all types of library oligomers, the basic oligonucleotide sequence can be shortened from both ends or split into two molecules to produce gaps or nicks in duplex DNA. Single nucleotides can be exchanged to add mismatches, substrate preferences, and so forth (Figures 1 and 2).
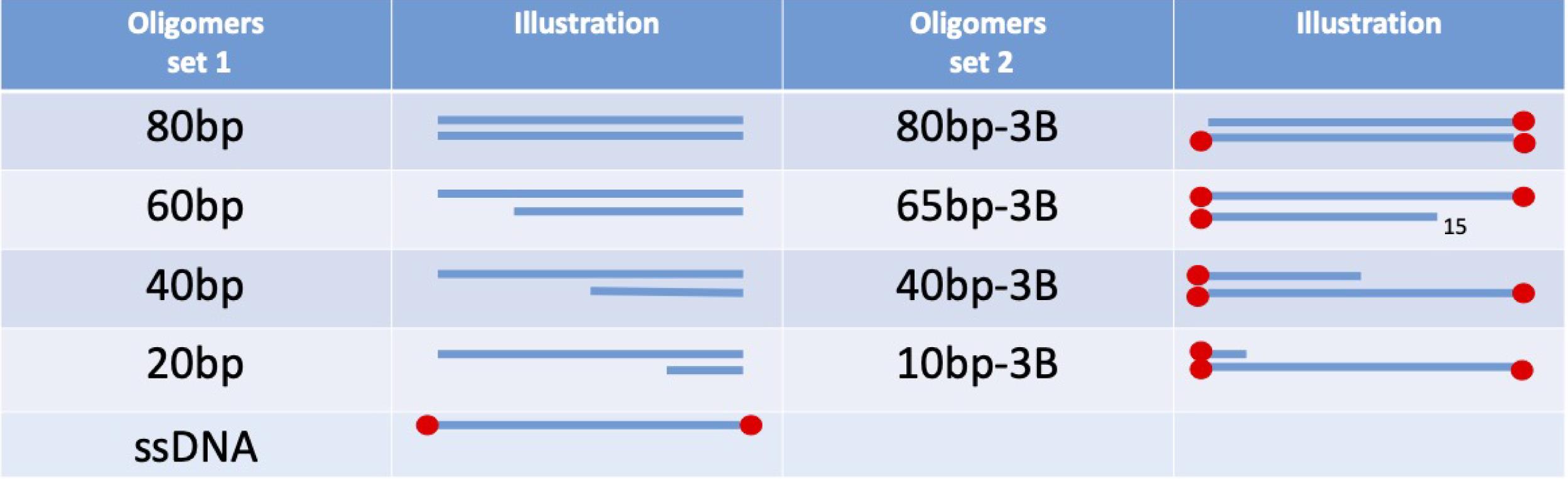
Figure 1. For the calibration curve, two example sets are presented, with and without streptavidin blockade on ends. These need to match the DNA substrates used in the experiment.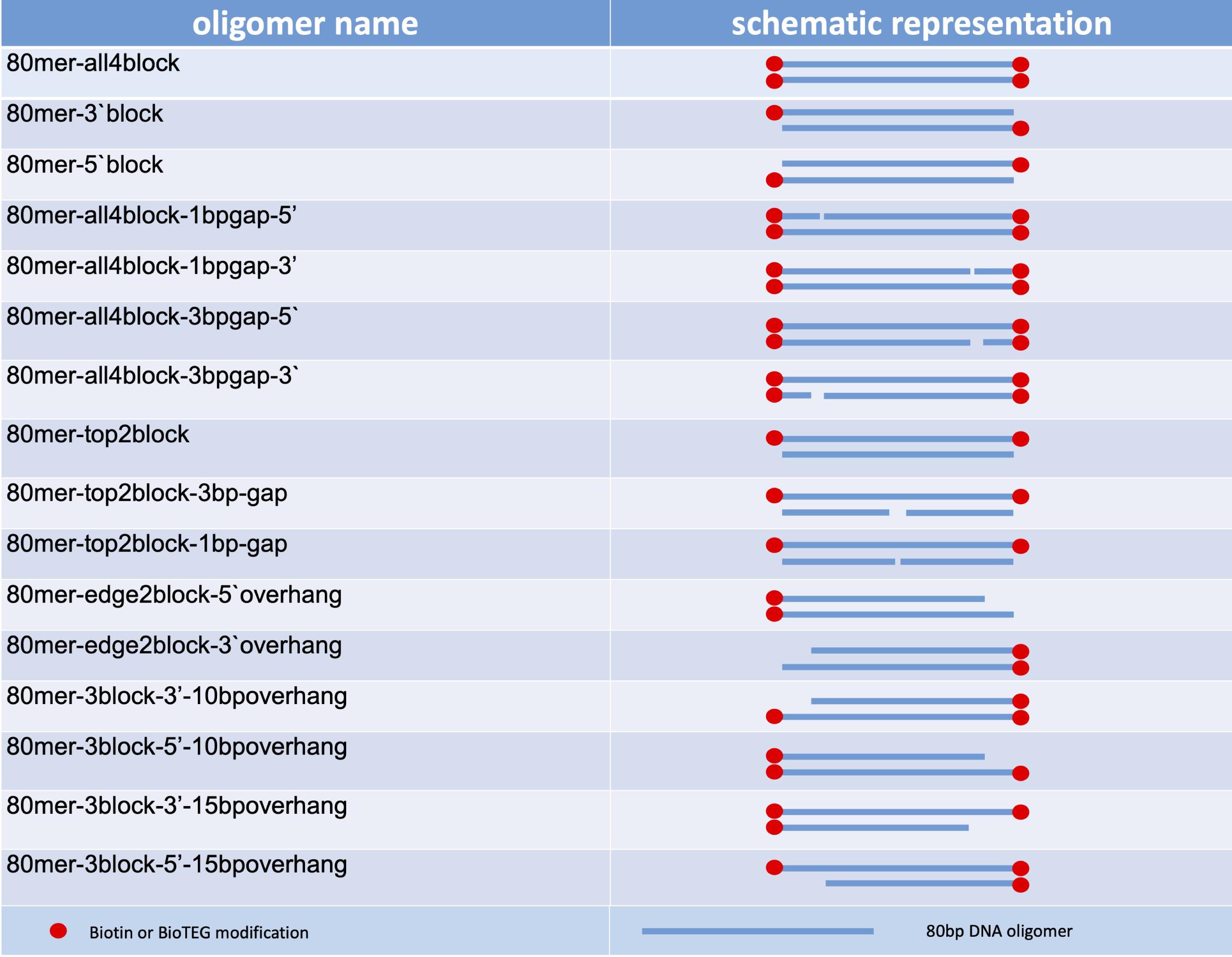
Figure 2. Set of 80-mer oligonucleotide substratesNucleases
Perform the calibration of the method using the commercial nucleases with known activity and preferably acting in a similar mechanism to the predicted activity of the enzyme to be tested. For example, to test the removal of 5′ mononucleotides from duplex DNA, use Exonuclease T7 (T7, New England Biolabs, catalog number: M0263S). For nucleases acting 3′ to 5′, use Exonuclease III (ExoIII, Thermo Scientific, catalog number: EN0191) (Figure 3).
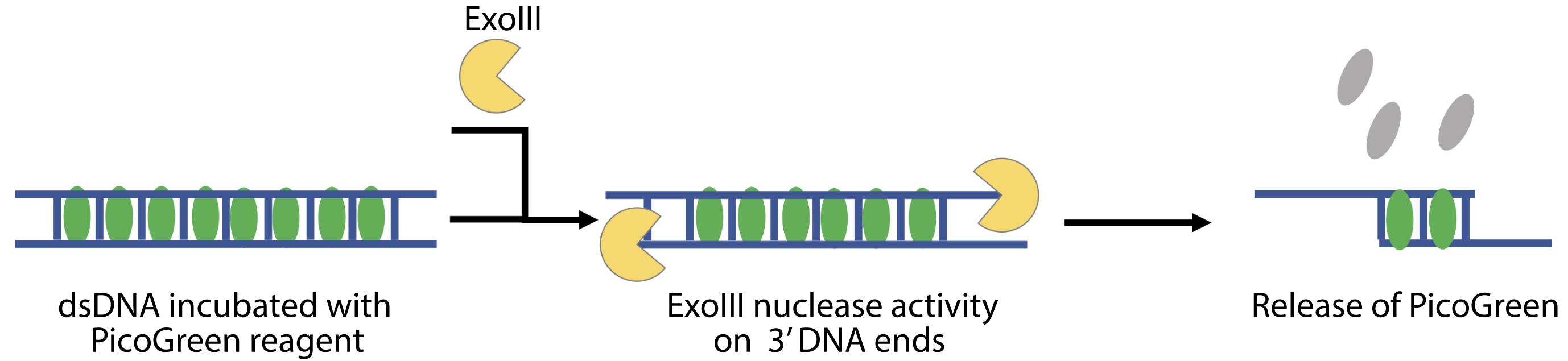
Figure 3. Scheme of PG release from dsDNA after nuclease treatment (ExoIII)1 M Tris, pH 7.5-8.0 (ThermoScientific, catalog number: 15567027)
5 M NaCl, RNase-free (ThermoScientific, catalog number: AM9759)
0.5 M EDTA pH 8.0 (ThermoScientific, catalog number: 15575020)
10× Tango Buffer (ThermoScientific, catalog number: BY5)
CutSmart (New England Biolabs, catalog number: B7204S)
Glycerol (ThermoScientific, catalog number: 15514011)
Quant-iTTM PicoGreenTM dsDNA Assay Kit (Invitrogen, catalog number: P7589)
Quant-iTTM microRNA Assay Kit (Invitrogen, catalog number: Q33140)
Annealing Buffer Composition (1×) (see Recipes)
Storage buffers (see Recipes)
Reaction buffers (see Recipes)
Divalent cations (see Recipes)
PG buffer (see Recipes)
Nucleic acid dyes (see Recipes)
Equipment
Plate reader (Tecan or BMG)
Select the appropriate measurement parameters (temperature, wavelength, number of flashes, and settle time) for your assay.
Preheat the microplate reader to the optimal temperature for enzyme activity (commonly 37°C). To slow down the reaction, set to a lower temperature (e.g., 20°C). Use the wavelength 483-15 nm for excitation (483 nm is the middle excitation peak with a bandwidth of 15 nm; i.e., the excitation range is 475-490 nm) and 530-30 nm for emission (530 nm is the middle emission peak with a bandwidth of 30 nm; i.e., the emission range is 515-545 nm). Increase the number of flashes per well until noise of BLANK wells does not improve further or until measurement time per well becomes unacceptable. Depending on the enzyme, read the samples every 45-60 s for 15 min to 2 h with a shake before each read.
Photobleaching occurring in the samples causes the fluorescence signal to decrease with time. Therefore, run a control curve run in every experiment simultaneously with samples to measure this effect. Longer readings can be inaccurate due to total photobleaching of PicoGreen to the level of the background.
We tested Infinite M Plex (TECAN) and CLARIOstar (BMG Labtech) plate readers. The nuclease assay can also be performed in the qPCR reader, but we found that the plate reader gives consistent results and offers more options, such as shaking the plate before a read.
While the best practices regarding reads per well vary with each enzyme, in general, an optimal duration of reads is approximately 1 h. If using a CLARIOstar, factor the whole time needed to read the entire plate, well by well, which for a full plate took approximately 48 s. So, each well could only be read every 50 s. If the plate reader permits it, try to use bidirectional reading row by row, and add the enzymes in the same order.
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Wyrzykowska, P., Rogers, S. and Chahwan, R. (2021). Measuring Real-time DNA/RNA Nuclease Activity through Fluorescence. Bio-protocol 11(21): e4206. DOI: 10.21769/BioProtoc.4206.
分类
癌症生物学 > 基因组不稳定性及突变 > 生物化学试验 > DNA结构和改变
癌症生物学 > 通用技术 > 生物化学试验
生物化学 > 蛋白质 > 活性
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link