- EN - English
- CN - 中文
Quantification of RuBisCO Expression and Photosynthetic Oxygen Evolution in Cyanobacteria
蓝藻RuBisCO表达和光合氧释放的定量研究
发布: 2021年10月20日第11卷第20期 DOI: 10.21769/BioProtoc.4199 浏览次数: 3210
评审: Alba BlesaRatnakar DeoleDhiraj Kumar Chaudhary

相关实验方案
将Miniprep制备的大肠杆菌K12菌株质粒DNA转化为可用于植物遗传转化的根癌农杆菌EHA105细胞的简单可靠方法
Beenzu Siamalube [...] Steven Runo
2025年01月05日 2178 阅读
Abstract
Phototrophic microorganisms are frequently engineered to regulate the expression and the activity of targeted enzymes of interest for specific biotechnological and agricultural applications. This protocol describes a method to evaluate the expression of RuBisCO (ribulose 1,5-bisphosphate carboxylase/oxygenase) in the model cyanobacterium Synechococcus elongatus PCC 7942, at both the transcript and protein levels by quantitative PCR and Western blot, respectively. We further describe an experimental method to determine photosynthetic activity using an oxygen electrode that measures the rate of molecular oxygen production by cyanobacterial cultures. Our protocol can be utilized to assess the effects of RuBisCO engineering at the metabolic and physiological levels.
Keywords: RuBisCO (RuBisCO)Background
RuBisCO (ribulose 1,5-bisphosphate carboxylase/oxygenase) is a central enzyme involved in fixation of atmospheric CO2 into biomass of its photoautotrophic producers (Nisbet et al., 2007; Tabita et al., 2008; Kacar et al., 2017; Erb and Zarzycki, 2018). Improving the efficiency of the RuBisCO carboxylation activity might increase the yield of produced biomass and drive cost-effective, ecologically friendly ways to biosynthesize diverse carbon-based compounds (Simkin et al., 2019; Kubis et al., 2019). Therefore, RuBisCO has become one of the most extensively studied and engineered enzymes (Bainbridge et al., 1995; Whitney et al., 2011). Different approaches have been utilized to manipulate the total activity of RuBisCO, such as improving its activation by activases (Bhat et al., 2017), incorporating beneficial mutations in the enzyme substrate-binding or active sites (Andersson, 2008), and upregulating RuBisCO expression (Parry et al., 2003; Carmo-Silva et al., 2015; Liang and Lindblad, 2017; Salesse-Smith et al., 2018; Durall et al., 2020; Garcia et al., 2021), as well genomically replacing the native RuBisCO with its ancient counterpart (Kedzior et al., 2021).
The effects of engineering strategies to increase the expression of RuBisCO are typically evaluated at both the transcript and protein levels. Here, we present a detailed protocol that builds and expands upon methods applied by others to analyze the expression of RuBisCO in cyanobacteria. The approach detailed here permits the assessment of the potential metabolic effect of altered enzyme expression by measuring the photosynthetic activity of engineered phototrophs. In particular, this protocol uses the model cyanobacterium Synechococcus elongatus PCC 7942 (hereafter S. elongatus) that is broadly employed in basic and applied research as it is naturally competent and its genome can easily be modified using standard engineering techniques (Atsumi et al., 2009; Taton et al., 2020). As an example of a modified strain that can be evaluated relative to wild-type S. elongatus (WT), we used a mutant strain “Syn02” that harbors the native rbc operon and a second copy inserted in the chromosome neutral site, as described and studied by our laboratory (Garcia et al., 2021).
Here, we have combined disparate pieces of operative methodological information in a single, readily available protocol. Furthermore, we adjusted methods reported elsewhere specifically to increase target yield and purity of isolated total RNA – required to generate cDNA templates used in qPCR analysis. Overall, our analysis of RuBisCO expression at the transcript level employs adapted protocols, including those recommended by different manufacturers (QIAGEN, Invitrogen, Applied Biosystems) to extract and purify total RNA from bacteria, reverse transcribe to cDNA, and analyze gene expression by qPCR. Specifically, we use reference gene secA for normalization of rbcL expression based on previous studies, showing its stable expression under diverse growth conditions in S. elongatus (Szekeres et al., 2014; Luo et al., 2019). To analyze expression at the protein level, total proteins were isolated from crude cell extracts under denaturing conditions, building on methods by Ivleva and Golden (2007) with modifications. The SDS-PAGE loading buffer containing 100 mM dithiothreitol and 2% (w/v) SDS is replaced with the hot Denaturing Lysis Buffer containing 1% (w/v) SDS without a reducing agent that would otherwise prohibit precise quantification of isolated proteins. The method for physical cell disruption by freeze-thaw cycles and glass beads is replaced with sonication. The procedure to perform Western blot is elaborated based on standard polyacrylamide gel electrophoresis and protein transfer protocols (Bio-Rad). Finally, directions for the use of a primary anti-RbcL antibody are adapted from Agrisera, and instructions on total protein stain and near-infrared secondary antibody-mediated immunodetection are provided by LI-COR Biosciences. Our protocol enables the isolation of ~5 mg/ml total protein. The procedure for oxygen evolution rate measurement follows manufacturer guidelines for the Oxygraph+ System (Hansatech Instruments) with some adjustments specific to cyanobacterial cultures based on previous methods (Liang and Lindblad, 2016; De Porcellinis et al., 2018). We additionally modified cyanobacteria preparation for the analysis (step E8 of this protocol) to decrease fluctuation during oxygen measurements and, thus, to improve reproducibility. Figure 1 illustrates all the major steps described in this protocol.
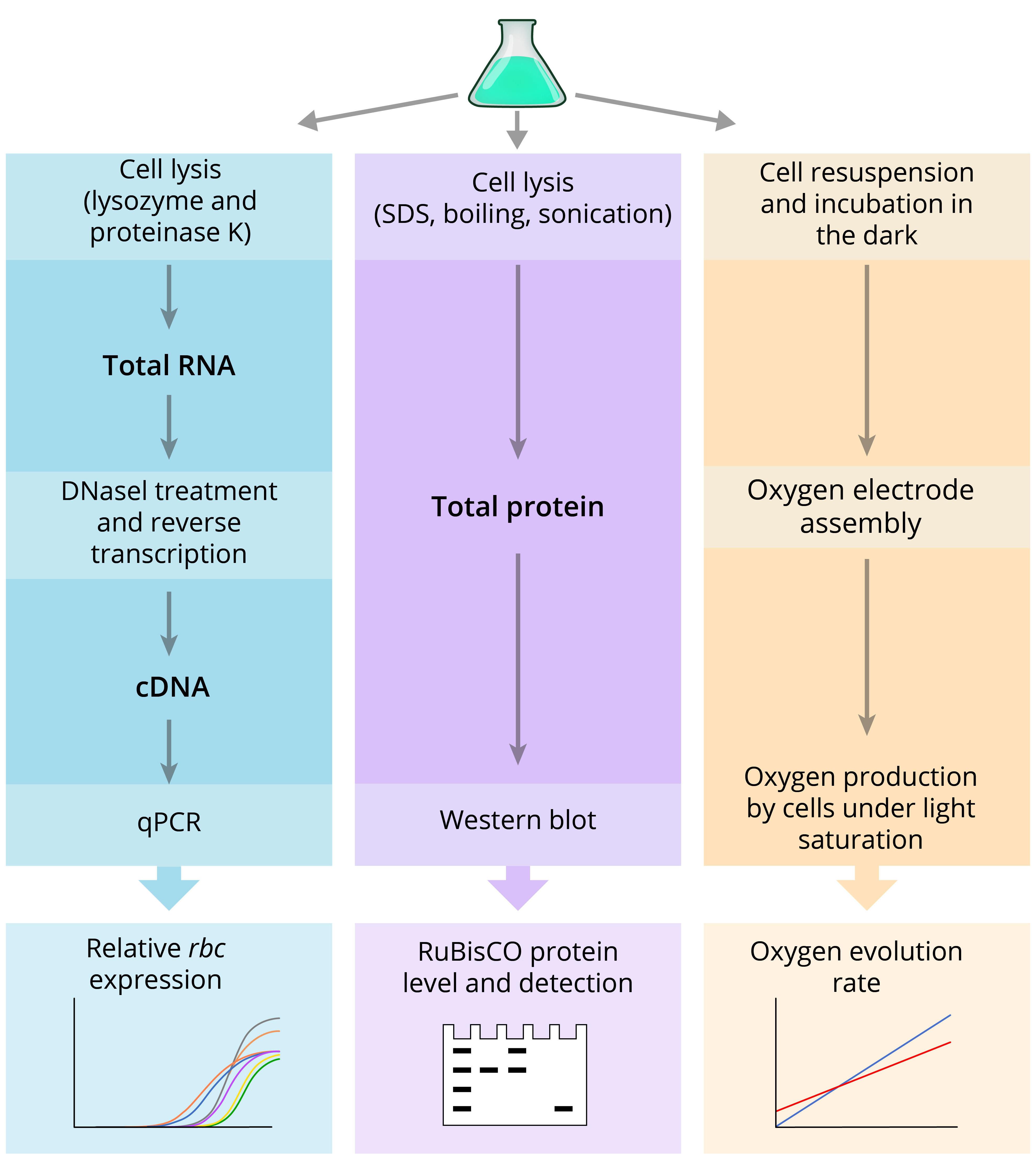
Figure 1. Schematic diagram of the experimental steps presented in this protocol
Materials and Reagents
96-well low-profile skirted PCR plates (Thermo Scientific, catalog number: AB0800W)
Synechococcus elongatus PCC 7942 WT (wild-type strain with one RuBisCO operon copy in the native site)
Synechococcus elongatus PCC 7942 Syn02 (engineered strain with two RuBisCO operon copies: one in the native site and the other in the neutral site; Garcia et al., 2021)
RNeasy Protect Bacteria Mini Kit (QIAGEN, catalog number: 74524)
Deoxyribonuclease I, amplification grade (Invitrogen, catalog number: 18068-015)
SuperScriptTM IV First-Strand Synthesis System (Invitrogen, catalog number: 18091050)
Q5® High-Fidelity DNA Polymerase (New England BioLabs, catalog number: M0491)
PowerUpTM SYBRTM Green Master Mix (Applied Biosystems, catalog number: A25742)
Agarose, LE, analytical grade (Promega, catalog number: V3125)
SYBRTM Safe DNA Gel Stain (Invitrogen, catalog number: S33102)
Bovine serum albumin (BSA) standards (Lee BioSolutions, catalog number: 100-10-0.25); concentrations [µg/ml]: 25, 125, 250, 500, 750, 1,000, 1,500, 2,000
PierceTM BCA Protein Assay Kit (Thermo Scientific, catalog number: 23225)
Tris (Thermo Fisher Scientific, catalog number: PRH5131)
Ammonium persulfate, APS (Sigma-Aldrich, catalog number: A3678)
TEMED (Bio-Rad, catalog number: 1610801)
30% acrylamide/bis solution (Bio-Rad, catalog number: 1610158)
Precision Plus ProteinTM Dual Color Standards (Bio-Rad, catalog number: 1610374)
RevertTM 700 Total Protein Stain (LI-COR Biosciences, catalog number: 926-11011)
Rabbit anti-RbcL antibody (Agrisera, catalog number: AS03 037)
Non-fat dry milk (LabScientific, catalog number: M0841)
IRDye® 800CW goat anti-rabbit IgG secondary antibody (LI-COR Biosciences, catalog number: 926-32211)
Potassium chloride (Sigma-Aldrich, catalog number: P9541)
Sodium dithionite/sodium hydrosulfite (Sigma-Aldrich, catalog number: 157953)
Methanol, anhydrous (Sigma-Aldrich, catalog number: 322415)
Sodium chloride (Sigma-Aldrich, catalog number: S9888)
Tween® 20 (Sigma-Aldrich, catalog number: P9416)
Ethylenediaminetetraacetic acid, EDTA (Sigma-Aldrich, catalog number: ED)
Lysozyme from chicken egg white (Sigma-Aldrich, catalog number: L4919)
Proteinase K (QIAGEN, catalog number: 19131)
Glycine (Thermo Fisher Scientific, catalog number: PRH5073)
Sodium dodecyl sulfate, SDS (Sigma-Aldrich, catalog number: L3771)
Acetic acid (Sigma-Aldrich, catalog number: 695092)
Intercept® (TBS) blocking buffer (LI-COR Biosciences, catalog number: 927-60001)
β-mercaptoethanol (Sigma-Aldrich, catalog number: M3148)
Ethanol, anhydrous (Sigma-Aldrich, catalog number: 443611)
Bromophenol blue (Sigma-Aldrich, catalog number: B0126)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Sodium nitrate, NaNO3 (Thermo Fisher Scientific, catalog number: BP360-500)
Magnesium sulfate heptahydrate, MgSO4·7H2O (Sigma-Aldrich, catalog number: 230391)
Calcium chloride dihydrate, CaCl2·2H2O (Sigma-Aldrich, catalog number: 223506)
Potassium phosphate dibasic, K2HPO4 (Sigma-Aldrich, catalog number: P8281)
Sodium carbonate, Na2CO3 (Supelco, catalog number: SX0395)
Citric acid monohydrate, C6H8O7·H2O (J.T. Baker, catalog number: 0115-01)
Ammonium iron(III) citrate, C6H11FeNO7 (Sigma-Aldrich, catalog number: F5879)
Ethylenediaminetetraacetic acid disodium salt, Na2EDTA·2H2O (Alfa Aesar, catalog number: A15161)
Boric acid, H3BO3 (J.T. Baker, catalog number: 0084-01)
Manganese(II) chloride tetrahydrate, MnCl2·4H2O (J.T. Baker, catalog number: 2540-01)
Zinc sulfate heptahydrate, ZnSO4·7H2O (Sigma-Aldrich, catalog number: Z0251)
Sodium molybdate dihydrate, Na2MoO4·2H2O (J.T. Baker, catalog number: 3764-01)
Copper(II) sulfate pentahydrate, CuSO4·5H2O (Sigma-Aldrich, catalog number: C8027)
Cobalt(II) nitrate hexahydrate, Co(NO3)2·6H2O (Sigma-Aldrich, catalog number: 239267)
BG-11 medium (see Recipes)
Denaturing Lysis Buffer (see Recipes)
Laemmli Sample Buffer (see Recipes)
Polyacrylamide resolving gel (see Recipes)
Polyacrylamide stacking gel (see Recipes)
Sodium dithionite saturated solution (see Recipes)
TAE Buffer (see Recipes)
TBS Buffer (see Recipes)
TBST Buffer (see Recipes)
TE Buffer (see Recipes)
TE Lysis Buffer (see Recipes)
TGS Buffer (see Recipes)
Transfer Buffer (see Recipes)
Equipment
250-ml glass flasks with sponge caps (Thermo Fisher Scientific)
Controlled environment chamber equipped with light source (Percival, model: I36LLVLC8)
UV-Vis Spectrophotometer (Agilent, model: Cary 60)
Heraeus Multifuge Centrifuge (Thermo Scientific, model: X3 FR)
Sorvall Legend Micro 17 Centrifuge (Thermo Scientific, catalog number: 75002431)
NanoDrop 2000c Spectrophotometer (Thermo Scientific, catalog number: ND-2000C)
Dry Bath Digital Heat Block (Benchmark, model: BSH1002)
Digital Shaker (VWR, model: 5000)
Analog Vortex Mixer (Fisher Scientific, model: 02215414)
Thermal Cycler (Applied Biosystems, model: 2720)
ABsolute qPCR plate seals (Thermo Scientific, catalog number: AB1170)
Real-Time Thermal Cycler (Analytik Jena AG, model: qTOWER3 G)
Microwave
250-ml beaker
Sonicator (Qsonica, model: Q125)
Nitrocellulose membrane (Thermo Scientific, catalog number: 88018)
Mini-PROTEAN® Tetra Vertical Electrophoresis Cell system (Bio-Rad, catalog number: 1658004)
Mini Trans-Blot Electrophoretic Transfer Cell system (Bio-Rad, catalog number: 1703930)
Odyssey® Fc Imaging System (LI-COR Biosciences, model: 2800-03)
Oxygraph+ System (Hansatech Instruments, catalog number: OXY1+)
Software
qPCRsoft (Analytik Jena AG, version: 3.1)
Image StudioTM (LI-COR Biosciences, version: 5.0)
Quantity One® 1-D Analysis (Bio-Rad, version: 4.6.7)
OxyTrace+ (Hansatech Instruments)
Microsoft 365 Excel
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Kędzior, M. and Kacar, B. (2021). Quantification of RuBisCO Expression and Photosynthetic Oxygen Evolution in Cyanobacteria. Bio-protocol 11(20): e4199. DOI: 10.21769/BioProtoc.4199.
分类
微生物学 > 微生物遗传学 > 基因表达
微生物学 > 微生物生理学 > 光合作用
生物科学 > 生物技术 > 微生物技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link