- EN - English
- CN - 中文
Analysis of Leukemia Cell Metabolism through Stable Isotope Tracing in Mice
小鼠白血病细胞代谢的稳定同位素示踪分析
发布: 2021年10月05日第11卷第19期 DOI: 10.21769/BioProtoc.4171 浏览次数: 5102
评审: Alak MannaNavnita DuttaAnonymous reviewer(s)
Abstract
Once thought to be a mere consequence of the state of a cell, intermediary metabolism is now recognized as a key regulator of mammalian cell fate and function. In addition, cell metabolism is often disturbed in malignancies such as cancer, and targeting metabolic pathways can provide new therapeutic options. Cell metabolism is mostly studied in cell cultures in vitro, using techniques such as metabolomics, stable isotope tracing, and biochemical assays. Increasing evidence however shows that the metabolic profile of cells is highly dependent on the microenvironment, and metabolic vulnerabilities identified in vitro do not always translate to in vivo settings. Here, we provide a detailed protocol on how to perform in vivo stable isotope tracing in leukemia cells in mice, focusing on glutamine metabolism in acute myeloid leukemia (AML) cells. This method allows studying the metabolic profile of leukemia cells in their native bone marrow niche.
Keywords: Cell metabolism (细胞代谢)Background
Unraveling the molecular regulation of cancer cells is critical to understand their insurgence, functioning, and potential therapeutic vulnerabilities. A dysregulated cellular metabolism is now widely accepted as a hallmark of many cancers (DeBerardinis and Chandel, 2016). Cancer cells display considerably different metabolic requirements compared to most normal differentiated cells and reprogram pathways controlling nutrient acquisition and metabolism to meet their specific bioenergetic, biosynthetic, and redox demands. Emerging evidence, mostly from solid tumors, shows that several metabolic characteristics of cancer cells can also be linked to therapeutic resistance (Zhao et al., 2013). Metabolic heterogeneity between cells, driven by genetic, cell state, or microenvironmental diversity, may further allow subpopulations of cells to escape therapy-related cell death (Martinez-Outschoorn et al., 2016).
The importance of examining the metabolism of cells in their native environment is underscored by several studies that have shown that artificial in vitro conditions can create metabolic dependencies that do not exist in vivo (Davidson et al., 2016; Cantor et al., 2017; Muir et al., 2017). However, studying metabolism in an in vivo setting brings several challenges. Gene or protein levels of metabolic enzymes may not always adequately reflect the metabolic state of a cell (Metallo and Vander Heiden, 2013). Measuring single metabolites in cells in vivo or on tissue sections is feasible, as is metabolic profiling of freshly-isolated cells through metabolomics analysis (Arnold et al., 2015; Faubert and DeBerardinis, 2017). These approaches have proven extremely valuable, but they provide a snapshot of metabolite levels and no dynamic information or insight into the flow of metabolites. Metabolic tracer analysis is a complementary method that uses stable isotope-labeled metabolites, which are taken up and metabolized by the cells, after which the fate of the labeled atoms (usually carbon, nitrogen, or hydrogen) can be analyzed by mass spectrometry. In this way, the relative activity of one or more selected metabolic pathways can be studied. Metabolic tracer analysis, as well as metabolic flux analysis, which combines metabolic tracer analysis with tracer uptake data and computational modeling, has become a standard technique in the study of cancer cell metabolism, offering insights into the relations between cell biology and biochemistry (Antoniewicz, 2018). While metabolic tracing is mainly performed with cells in culture, it is also more and more used in in vivo settings, both in animal models and in humans (Faubert and DeBerardinis, 2017; Fernández -García et al., 2020). Most in vivo metabolic tracing studies have focused on solid tumors, analyzing the tumor as a whole, without separating the cancer cells from other cells of the tumor microenvironment, such as immune cells, stromal cells, or blood vessel cells. The advantage of working with liquid tumors is that the cancer cells can be readily obtained from the bone marrow without the need for enzymatic tissue digestion and can be separated from the other cells by fluorescence-activated cell sorting (FACS), thus allowing specific analysis of cancer cell metabolism in vivo.
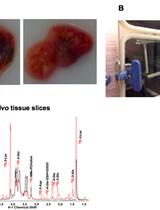
In our original study, we investigated the metabolic adaptations of acute myeloid leukemia (AML) cells in vivo during the course of induction chemotherapy treatment and identified transient changes in glutamine levels (van Gastel et al., 2020). We performed in vivo stable isotope tracing, according to the protocol described here, to understand the metabolic fate of glutamine in control AML cells or cells persisting after chemotherapy. We used a transplant-based mouse model of human AML, driven by the MLL-AF9 fusion protein (Corral et al., 1996). The AML cells also expressed GFP, which greatly facilitates cell sorting. The protocol described below should be compatible with any mouse leukemia model in which the leukemia cells express a fluorescent marker. It is also possible to perform metabolic tracing using our protocol in other hematopoietic cell populations, such as normal hematopoietic progenitor cells (Lineage-cKit+), as long as they can be readily isolated from bone marrow and separated from other cells by FACS.
Materials and Reagents
5 ml round bottom polystyrene test tube with cell strainer snap cap (Corning, Falcon, catalog number: 352235)
50 ml centrifuge tube, conical bottom (Corning, Falcon, catalog number: 352070)
40 µm cell strainer (Corning, Falcon, catalog number: 352340)
300 µl insulin syringe with 31 G needle (BD Biosciences, catalog number: 328440)
1 ml syringe (VWR, catalog number: 612-0106)
Needle, 25 G × 5/8 (BD Biosciences, catalog number: 305122)
K2-EDTA Microtainer tubes (BD Biosciences, catalog number: 365974)
1.7 ml Posi-Click microcentrifuge tubes (Denville, catalog number: C-2170)
Alcohol preparatory pads (Covidien, WEBCOL, catalog number: 89029-964)
LC/MS sample vials (ThermoFisher, catalog number: C4000-11)
LC/MS vial caps (ThermoFisher, catalog number: C5000-54B)
ZORBAX Extend-C18, 2.1 × 150 mm, 1.8 μm (Agilent, catalog number: 759700-902)
L-glutamine, 13C5, 99% (Cambridge Isotope Laboratories, catalog number: CLM-1822-H)
L-glutamine, 15N2, 98% (Cambridge Isotope Laboratories, catalog number: NLM-1328)
Sodium chloride (Sigma-Aldrich, catalog number: S5886)
Water for UHPLC, for mass spectrometry (Sigma-Aldrich, catalog number: 900682)
Methanol for UHPLC, for mass spectrometry (Sigma-Aldrich, catalog number: 900688)
Dulbecco’s phosphate-buffered saline (PBS), without calcium and magnesium (Corning, catalog number: 21-031-CV)
Fetal bovine serum (FBS), USDA tested (Cytiva, HyClone, catalog number: SH3091003)
ACK lysing buffer (ThermoFisher, Gibco, catalog number: A1049201)
7-AAD (BD Biosciences, BD Pharmingen, catalog number: 559925)
Tributylamine, for liquid chromatography (Fisher Scientific, catalog number: 60-046-943)
Glacial acetic acid, for liquid chromatography (Fisher Scientific, catalog number: A35-500)
Saline (see Recipes)
Flow buffer (see Recipes)
Cell lysis buffer (see Recipes)
Mobile Phase Buffer A (see Recipes)
Mobile Phase Buffer B (see Recipes)
Equipment
Mouse restraining device (Braintree Scientific Inc., Tailveiner Restrainer, catalog number: TV-150 STD)
Micro dissecting scissors (Roboz Surgical, catalog number: RS-5906SC)
Narrow pattern forceps (Fine Science Tools, catalog number: 11002-12)
Mortar (VWR, catalog number: 89038-144)
Pestle (VWR, catalog number: 89038-160)
Refrigerated swinging bucket centrifuge 5910R with S-4xUniversal rotor (Eppendorf, catalog number: 5942000315)
Refrigerated microcentrifuge 5430R with FA-45-30-11 rotor (Eppendorf, catalog number: 5428000010)
Analytical balance (Mettler Toledo, catalog number: ME54)
BD FACSAria II Cell Sorter (BD Biosciences)
SpeedVac Vacuum Concentrator (ThermoFisher, catalog number: RVT450)
LP Vortex Mixer (ThermoFisher, catalog number: 11676331)
6470 Triple Quadrupole Mass Spectrometer (Agilent)
Software
BD FACSDiva Software, version 8.0.2 (BD Biosciences)
Excel (Microsoft)
Mass Hunter, version B.08.00 (Agilent)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
van Gastel, N., Spinelli, J. B., Haigis, M. C. and Scadden, D. T. (2021). Analysis of Leukemia Cell Metabolism through Stable Isotope Tracing in Mice. Bio-protocol 11(19): e4171. DOI: 10.21769/BioProtoc.4171.
分类
癌症生物学 > 细胞能量学 > 肿瘤微环境 > 新陈代谢
癌症生物学 > 癌症生物化学 > 癌代谢
细胞生物学 > 细胞新陈代谢 > 氨基酸
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link