- EN - English
- CN - 中文
A Novel Method to Map Small RNAs with High Resolution
一种新的高分辨率小RNA图谱绘制方法
发布: 2021年08月20日第11卷第16期 DOI: 10.21769/BioProtoc.4128 浏览次数: 4678
评审: Alessandro DidonnaIgnacio LescanoAnonymous reviewer(s)
Abstract
Analyzing cellular structures and the relative location of molecules is essential for addressing biological questions. Super-resolution microscopy techniques that bypass the light diffraction limit have become increasingly popular to study cellular molecule dynamics in situ. However, the application of super-resolution imaging techniques to detect small RNAs (sRNAs) is limited by the choice of proper fluorophores, autofluorescence of samples, and failure to multiplex. Here, we describe an sRNA-PAINT protocol for the detection of sRNAs at nanometer resolution. The method combines the specificity of locked nucleic acid probes and the low background, precise quantitation, and multiplexable characteristics of DNA Point Accumulation for Imaging in Nanoscale Topography (DNA-PAINT). Using this method, we successfully located sRNA targets that are important for development in maize anthers at sub-20 nm resolution and quantitated their exact copy numbers.
Graphic abstract:
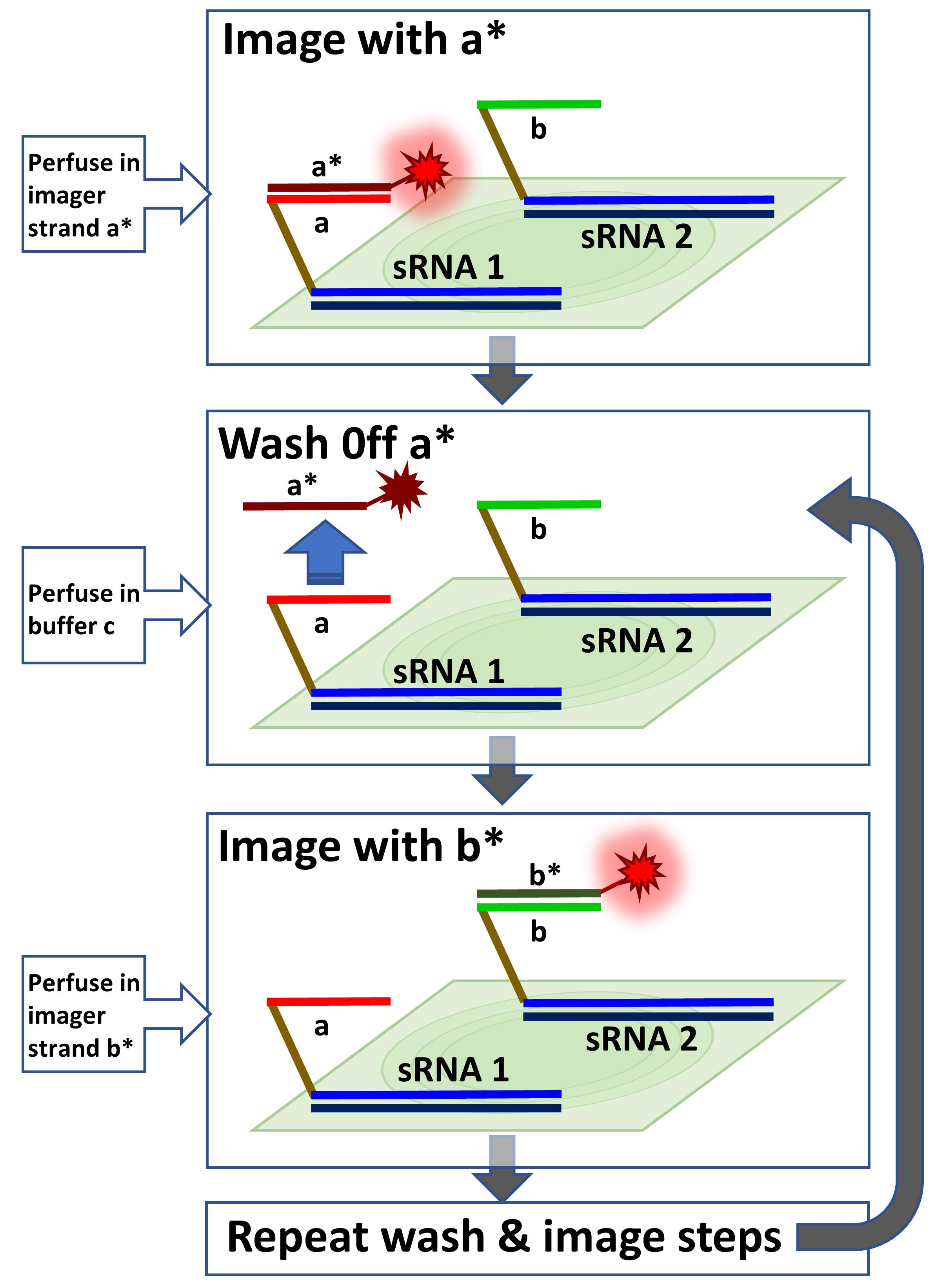
Multiplexed sRNA-PAINT. Multiple Vetting and Analysis of RNA for In Situ Hybridization (VARNISH) probes with different docking strands (i.e., a, b, …) will be hybridized to samples. The first probe will be imaged with the a* imager. The a* imager will be washed off with buffer C, and then the sample will be imaged with b* imager. The wash and image steps can be repeated sequentially for multiplexing.
Background
Light diffraction limits restrict imaging fluorophores closer than ~200 nm (Hell, 2009). Several imaging technologies have been created to overcome this limitation by exploiting transient fluorophore ON and OFF states (blinking), including stimulated emission depletion (STED) (Blom and Widengren, 2017), photoactivated localization microscopy (PALM) (Shroff et al., 2008), structured illumination microscopy (SIM) (Heintzmann and Huser, 2017), and stochastic optical reconstruction microscopy (STORM) (Bates et al., 2013). STORM increases the imaging resolution to around 30 nm (Xu and Liu, 2019); however, it relies on the stochastic blinking of fluorophores. As a result, multiplexing is often limited by the choice of fluorophores with ideal photophysical properties, including high photon numbers, low duty cycles, and high survival fractions, such as Alexa Fluor 647 and Dyomics 654 (Dempsey et al., 2011). Moreover, image quantitation is often challenging due to the unpredictability of the blinking properties of the chosen fluorophores and photobleaching (Schnitzbauer et al., 2017; Huang et al., 2020).
DNA Point Accumulation for Imaging in Nanoscale Topography (DNA-PAINT) was invented for ultra-high, sub-5 nm spatial detection of target protein molecules (Jungmann et al., 2014; Schnitzbauer et al., 2017; Strauss and Jungmann, 2020). It utilizes the base-pairing nature of single-stranded DNA molecules to their reverse complement strands. In the DNA-PAINT design, an approximately 10 nucleotide (nt)-long docking strand is linked to a target molecule via a biotin-streptavidin bridge or DBCO-sulfo-NHS ester chemical reaction (Schnitzbauer et al., 2017). An imager strand, conjugated to a fluorophore, is perfused in during the imaging process. The melting temperature (Tm) of the imager and docking strands is designed to be ~15°C. As a result, the imager strand-docking strand pairs will constantly bind and unbind at room temperature, which contributes to the blinking events, similar to the other super-resolution microscopy imaging techniques. The predictability of the binding duration and unlimited docking-imager strand combinations make DNA-PAINT ideal for quantitation and multiplexing (Jungmann et al., 2016).
sRNA-PAINT was created based on DNA-PAINT to detect small RNAs (sRNAs) that are usually ~10 nm in size (Huang et al., 2020). Our method preserves the nanometer resolution, quantitation, and multiplexing advantages of DNA-PAINT. In addition, we have introduced efficient and specific detection of sRNA targets with locked nucleic acid (LNA) (McTigue et al., 2004) and probe immobilization with 1-ethyl-1-(3-dimethylaminopropyl) carbodiimide (EDC) (Pena et al., 2009). sRNA-PAINT can be used for the multiplexed detection of sRNAs, their precursor mRNAs, and proteins in the sRNA biogenesis pathway when employed in combination with DNA-PAINT for understanding the relative localization patterns of small RNAs and their biogenesis machinery.
Materials and Reagents
20-ml glass scintillation vials (Electron Microscopy Sciences, catalog number: 72632)
50-ml Falcon tubes (Thermo Fisher Scientific, Corning, catalog number: 14-432-22)
0.5 M ethylenediaminetetraacetic acid (EDTA) solution (pH 8.0) (Thermo Fisher Scientific, Fisher BioReagentsTM, catalog number: BP2482-500)
1 M Tris-HCl solution (pH 8.0) (Thermo Fisher Scientific, catalog number: AAJ22638AP)
16% paraformaldehyde (Electron Microscopy Sciences, catalog number: RT15710) (store at 4°C)
1-ethyl-3- (3-dimethylaminopropyl) carbodiimide (EDC) (store at -20°C with a nitrogen seal)
200 proof ethanol (Thermo Fisher Scientific, Fisher BioReagentsTM, catalog number: 64-17-5) (store in a flammable cabinet)
Deionized formamide (Millipore Sigma, catalog number: S4117) (store at 4°C)
Denhardt’s solution (50×) (Millipore Sigma, catalog number: D2532) (store at -20°C)
Dextran sulfate sodium salt (Millipore Sigma, catalog number: 67578) (store at 4°C)
Histoclear II (VWR, Electron Microscopy Sciences, catalog number: 101412-878)
NaCl (Millipore Sigma, catalog number: NIST975A)
Paraffin wax pellets (Electron Microscopy Sciences, Paraplast X-tra, catalog number: 19214)
Saline-sodium citrate (SSC) buffer (20×) (Thermo Fisher Scientific, Fisher BioReagentsTM, catalog number: BP1325)
Sodium phosphate (Millipore Sigma, catalog number: 324283)
tRNA (Millipore Sigma, catalog number: R1753) (store at -20°C)
Ethanol gradient solutions (see Recipes)
PHEM buffer (2×) (see Recipes)
Fixation buffer (see Recipes)
Enzyme solution (see Recipes)
Glycine solution (see Recipes)
TE buffer (see Recipes)
TE-protease solution (see Recipes)
Dextran sulfate solution (50%) (see Recipes)
Highly abundant sRNA (see Recipes)
Hybridization salt (see Recipes)
Hybridization buffer (see Recipes)
TBS buffer (pH 7.5) (see Recipes)
tRNA solution (100 mg/ml) (see Recipes)
Buffer C (see Recipes)
EDC buffer (see Recipes)
EDC solution (see Recipes)
Equipment
DH40iL culture dish incubation system (Warner Instruments, model: 640388)
Dissecting microscope (Zeiss M2BIO Dissecting Microscope)
Flat-bottomed containers (Pyrex storage, 6 cup-1.5L)
Fluigent Aria perfusion (Fluigent, Parts no: CB_SY_AR_1)
Forceps (Electron Microscopy Sciences, catalog number: 72997-20)
Glass staining dishes (Electron Microscopy Sciences, catalog number: 71426-DL)
Grace Bio-Labs HybriSlip hybridization covers (Millipore Sigma, catalog number: 726024)
Heat incubator (Fisher Scientific Isotemp incubator)
Heat block for 1.5-ml tubes (BioExpress, Mini Dry Bath, model number: AS-BSH200-471)
Hybridization oven (UVP HB-1000 hybridizer, model number: 95-0030-01)
Masterflex C/L peristaltic pump (60 RPM) (Cole-Parmer Instrument, model: 77120-62)
Paraffin microtome (Microm, model: MRK0403643)
Parafilm (Thermo Fisher Scientific, BemisTM, catalog number: PM998)
Plastic wrap (Thermo Fisher Scientific, FisherbrandTM Clear Plastic Wrap, catalog number: 12-640)
Quick-release magnetic chamber for 25-mm low-profile round coverslips (Warner Instruments, model: 641943)
Razor blades (Thermo Fisher Scientific, FisherbrandTM Razor Blades, catalog number: 22-305654)
Slide warmer (Thermo Fisher Scientific, FisherbrandTM Slide Drying Bench, catalog number: 11-474-470)
Stainless-steel glass rack (Electron Microscopy Sciences, catalog number:71426-R)
Superfrost glass slides (Thermo Fisher Scientific, FisherbrandTM Tissue Path SuperfrostTM Plus Gold Slides, catalog number: 15-188-48)
Tissue embedding and processing cassettes (Electron Microscopy Sciences, catalog number: 70070)
Vacuum bell jar (Jelo Tech, model number: F42400-2221 with a Nisshin vacuum gauge, model number: B53595)
Vacuum pump (Welch, model number: K48ZZMEM31)
ValveLink8.2 Perfusion System (AutoMate Scientific, Berkeley, CA)
Watercolor paintbrushes (#0)
Wide Spectral Band 600 ± 100 nm Gold Fiducials coverglass (600-100AuF; Hestzig LLC, Leesburg, VA)
Single-molecule localization microscope: Zeiss Elyra PS.1 super-resolution microscope (Carl Zeiss, Lberkochen, Germany) or Andor Dragonfly (Oxford Instruments, Abingdon, United Kingdom).
Software
ImageJ ThunderSTORM (https://zitmen.github.io/thunderstorm/) (Ovesny et al., 2014)
Picasso (https://github.com/jungmannlab/picasso) (Schnitzbauer et al., 2017)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Huang, K., Demirci, F., Meyers, B. C. and Caplan, J. L. (2021). A Novel Method to Map Small RNAs with High Resolution. Bio-protocol 11(16): e4128. DOI: 10.21769/BioProtoc.4128.
分类
植物科学 > 植物分子生物学 > RNA > RNA 检测
植物科学 > 植物细胞生物学 > 细胞成像
细胞生物学 > 细胞成像 > 超分辨率成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link