- EN - English
- CN - 中文
A Gel-Based Assay for Probing Protein Translocation on dsDNA
一种基于凝胶的检测dsDNA蛋白易位的方法
发布: 2021年07月20日第11卷第14期 DOI: 10.21769/BioProtoc.4094 浏览次数: 4333
评审: Emilia KrypotouLaura Molina-García
Abstract
Protein translocation on DNA represents the key biochemical activity of ssDNA translocases (aka helicases) and dsDNA translocases such as chromatin remodelers. Translocation depends on DNA binding but is a distinct process as it typically involves multiple DNA binding states, which are usually dependent on nucleotide binding/hydrolysis and are characterized by different affinities for the DNA. Several translocation assays have been described to distinguish between these two modes of action, simple binding as opposed to directional movement on dsDNA. Perhaps the most widely used is the triplex-forming oligonucleotide displacement assay. Traditionally, this assay relies on the formation of a DNA triplex from a dsDNA segment and a short radioactively labeled oligonucleotide. Upon translocation of the protein of interest along the DNA substrate, the third DNA strand is destabilized and eventually released off the DNA duplex. This process can be visualized and quantitated by polyacrylamide electrophoresis. Here, we present an effective, sensitive, and convenient variation of this assay that utilizes a fluorescently labeled oligonucleotide, eliminating the need to radioactively label DNA. In short, our protocol provides a safe and user-friendly alternative.
Graphical abstract:
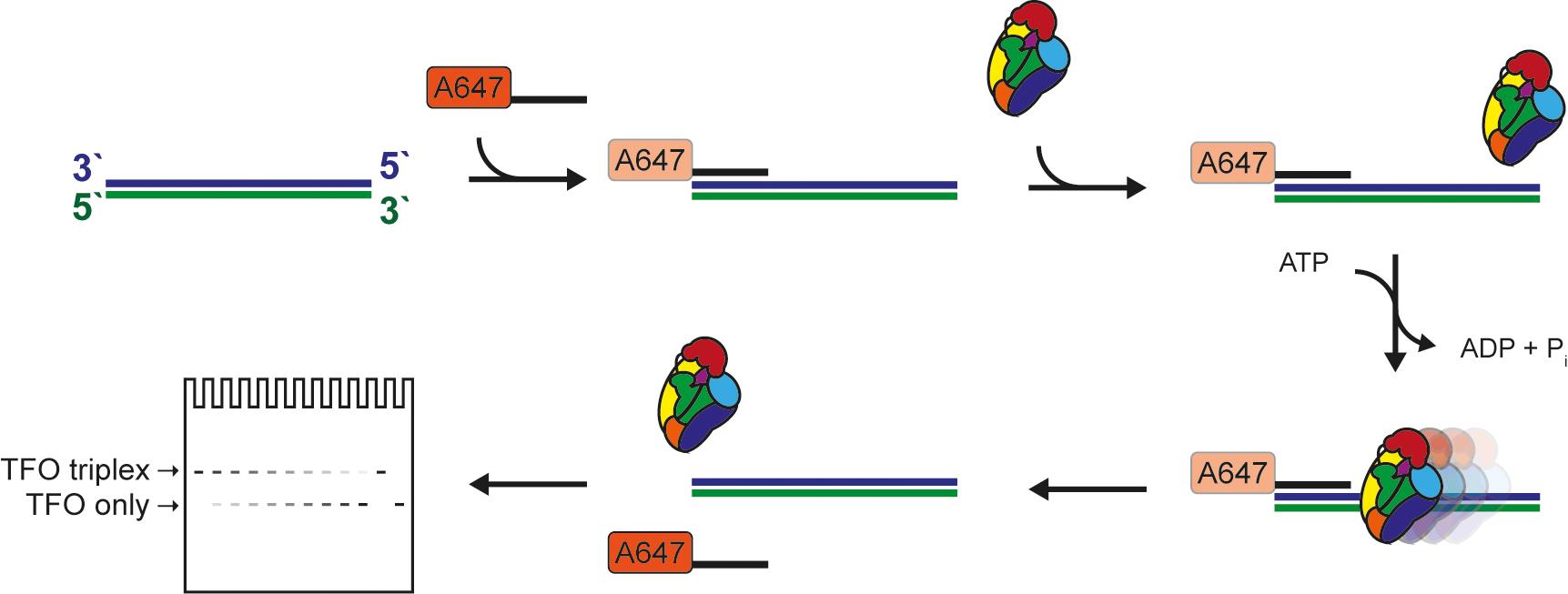
Figure 1. Schematic of the triplex-forming oligonucleotide displacement assay.
Background
DNA translocases and DNA helicases are structurally related and typically contain an ATP-binding core composed of two RecA-type lobes (Singleton et al., 2007) connected to accessory and family-specific domains that modulate the activity of the motor. While DNA helicases have the ability to processively move along single-stranded DNA or RNA, thereby unwinding the two strands, dsDNA translocases cannot, as these move on dsDNA instead with varied processivities. Altogether, ss/ds-DNA tracking ATPases are clustered into superfamilies (SF1-SF6) depending on their translocation mechanism, domain organization, and oligomerization state (Brosh and Matson 2020). Translocation requires multiple steps – at least two – first binding of the protein to the nucleic acid (loading) and second, sliding along it. The role of nucleotide in these processes is very much family-specific and does not always require ATP hydrolysis (Fairman-Williams et al., 2010). Therefore, translocases can have varied processivities and can operate using an active, inchworming mechanism or as Brownian motors (Briggs and Fischer 2014). Crystal structures of translocases mostly belonging to SF1 and SF2 (Subramanya et al., 1996; Korolev et al., 1997; Velankar et al., 1999; Tomko et al., 2007; Gu and Rice 2010), and, more recently, electron microscopy studies of chromatin remodelers (Yan et al., 2019; Han et al., 2020; Wagner et al., 2020), have contributed insightful mechanistic details. However, one must bear in mind that these provide still images of isolated steps in the catalytic cycle and rarely the complete trajectory of states underlying translocation.
Unlike assays for helicase activity, assays to monitor translocation on dsDNA are more challenging for the simple fact that no easily quantifiable product is formed. Some dsDNA translocases, such as select chromatin remodelers, have been tested in “cruciform extrusion” assays quantifying changes in superhelicity, which can be correlated with translocation (Havas et al., 2000). However, the most popular assay for dsDNA translocation remains, by far, the triplex-forming oligonucleotide (TFO) displacement assay (Firman and Szczelkun 2000).
As a general principle, a DNA triplex is formed by annealing a short TFO to a dsDNA substrate with a TFO binding site (TBS), and the structure is held together via Hoogsteen base-pairing (Gowers and Fox 1999). Triplex formation requires a low pH environment. Once the triplex is formed, it is no longer pH dependent, and the assay can be performed at neutral pH (Kopel et al., 1996) if Mg2+ is present (Singleton and Dervan 1993). The substrate can differ in length, depending on the protein to be tested and its processivity (if known), and the TFO is either labeled with 32P or a fluorophore. The translocase will bind preferentially or exclusively to the double-stranded portion of the substrate, move along the DNA, and eventually dislodge the weakly bound TFO, which can then be detected. There are caveats to be considered when labeling the TFO either radioactively or fluorescently. In the former case, safety precautions regarding work with, storage, and disposal of radioactive materials have to be considered. On the other hand, fluorophores can interact non-specifically with proteins. Radioactivity-based assays are end-point and discontinuous based on detection of bound and free TFO on agarose or polyacrylamide gels (Firman and Szczelkun 2000; Smith et al., 2007). In contrast, the use of a fluorescently-labeled TFO can be coupled with either a gel-based end-point or continuous monitoring assays. The continuous assay exploits the attribute of fluorophores to change the intensity of the fluorescent signal when bound by the DNA duplex. In short, the signal will be “brighter” for the free TFO and “dimmer” when the TFO is incorporated within the triplex (McClelland et al., 2005). Although using a continuous assay has the advantage of monitoring translocation at every step, it requires access to a stop-flow fluorimeter.
In our assay, we combined elements of both approaches, e.g., the use of a fluorescently-labeled TFO and the ease of quantitation based on electrophoresis through standard commercially available gels. This makes this assay accessible for the majority of biochemical labs.
Special Considerations When Designing a TFO Displacement Assay
The key requirement for designing a successful TFO displacement assay is the sequence of the TFO – a single mismatch between TFO and dsDNA substrate can be destabilizing (Puri et al., 2004). Often, the following TFO sequence is used: TTTCTTTCTTCTTTCTTTTCTT. Next, the assembly of the triplex DNA has to be performed under low pH conditions (Maher et al., 1990; Plum et al., 1990). During the displacement assay, the pH can be raised to neutral as long as Mg2+ is present in the reaction buffer (Singleton and Dervan 1993). Depending on the protein of interest, the length of the dsDNA substrate can differ between short (in the range of 50 bp) and long (in the kb range). If the studied protein recognizes a specific DNA motif, it can be integrated into the substrate. Protein concentration is important, and 100-500 nM is a good starting point, but high protein concentrations can interfere with translocation. The assay can be performed at a range of temperatures, but temperatures above 40°C lead to significant spontaneous dissociation of the DNA triplex, and the timeframe of the experiment has to be adjusted. In case of contaminating DNAses, an unlabeled single-stranded oligonucleotide can be added to act as a nuclease trap as long as this has negligible affinity for the DNA translocase of interest
Materials and Reagents
Eppendorf tubes (Denville Scientific, catalog number: C2170; store at RT)
Opaque tubes (Argos Technologies, catalog number: T7456-001; store at RT)
PCR tubes (Corning Thermowell Gold PCR 0.2 ml, 1×8 strips, catalog number: 3741; store at RT)
Protein of interest
Tris Base (Fisher Bioreagents, catalog number: BP152-1; store at RT)
MES (2-(N-Morpholino)ethanesulphonic acid monohydrate) (Alfa Aesar, catalog number: A16104-22; store at RT)
MOPS (3-(N-Morpholino)propanesulfonic acid) (Sigma Aldrich, catalog number: M3183-500g; store at RT)
HCl (Hydrochloric acid) (Fisher Chemical, catalog number: A144-500; store at RT)
Acetic acid (Fisher Chemical, catalog number: A38-212; store at RT)
NaCl (Fisher Scientific, catalog number: S271-500; store at RT)
MgCl2·6H2O (Fisher Scientific, catalog number: M33-500; store at RT)
Glucose (Sigma Aldrich, catalog number: G8270-1kg; store at RT)
SDS (Fisher BioReagents, catalogue number: BP8200500; store at RT)
EDTA (Ethylenediaminetetraacetic acid) (Sigma Aldrich, catalog number: EDS-500g; store at RT)
DTT (Dithiothreitol) (Gold Biotechnology, catalog number: DTT50; store at -20°C)
Beta-mercaptoethanol (Arcos Organics, catalog number: AC125472500; store at RT)
NuPAGE 3-8%Tris Acetate Protein Gels, 1-0 mm, 12 well (Thermo Fisher Scientific, catalog number: EA03752BOX; store at 4°C)
Alexa-647 labeled TFO oligonucleotide (Invitrogen; store at -20°C in opaque tubes, avoid repeated thaw-freeze cycles, TTTCTTTCTTCTTTCTTTTCTT)
Single-stranded DNA substrate with TFO binding site (TBS) (Invitrogen; store at -80°C, top and bottom strand)
Short single-stranded DNA oligonucleotide (Invitrogen; store at -20°C)
ATP (Roche Applied Science, catalog number: 10519987001; store at -80°C)
Running buffer (see Recipes)
GSM buffer (see Recipes)
Reaction buffer (see Recipes)
Dialysis buffer (see Recipes)
Triplex formation buffer (see Recipes)
Annealing buffer (see Recipes)
1× TE buffer (see Recipes)
Equipment
Power Source (BioRad, model: Powerpac HC 1645052)
Novex Bolt Mini Gel Tank (Invitrogen, model: A25977)
Heat block (American Scientific Products, model: H2025-1)
PCR machine (Eppendorf, model: Mastercycler EP Gradient Model 5341)
ChemiDoc MP (BioRad, model: 12003154)
Software
Image Lab (BioRad, https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z) or equivalent software like ImageJ (https://imagej.nih.gov/ij/)
Microsoft Excel (Microsoft, https://www.microsoft.com/en-us/microsoft-365/excel)
GraphPad Prism 7 (GraphPad, https://www.graphpad.com/scientific-software/prism/)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Brugger, C. and Deaconescu, A. M. (2021). A Gel-Based Assay for Probing Protein Translocation on dsDNA. Bio-protocol 11(14): e4094. DOI: 10.21769/BioProtoc.4094.
分类
生物化学 > 蛋白质 > 相互作用 > 蛋白质-DNA相互作用
分子生物学 > DNA > DNA-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link