- EN - English
- CN - 中文
CRISPR/Cas9-mediated Precise SNP Editing in Human iPSC Lines
CRISPR/Cas9介导的人iPSC细胞系SNP精确编辑
发布: 2021年06月20日第11卷第12期 DOI: 10.21769/BioProtoc.4051 浏览次数: 6305
评审: Jaira Ferreira de VasconcellosGiovanna PiovaniAnonymous reviewer(s)

相关实验方案
利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1231 阅读
Abstract
Human induced pluripotent stem cells (hiPSCs) have been extensively used in the fields of developmental biology and disease modeling. CRISPR/Cas9 gene editing in iPSC lines often has a low frequency, which hampers its application in precise allele editing of disease-associated single nucleotide polymorphisms (SNPs), especially those in the noncoding parts of the genome. Here, we present a unique workflow to engineer isogenic iPSC lines by SNP editing from heterozygous to homozygous for disease risk alleles or non-risk alleles using a transient and straightforward transfection-based protocol. This protocol enables us to simultaneously obtain pure and clonal isogenic lines of all three possible genotypes of a SNP site within about 4 to 5 weeks.
Keywords: Induced pluripotent stem cells (诱导多能干细胞)Background
The application of CRISPR/Cas9 gene editing to iPSC cells provides unrivalled potential in the fields of developmental biology, disease modeling, and regenerative medicine. However, since iPSCs are not especially acquiescent to the traditional strategies used in CRISPR/Cas9-gene editing, the editing efficiency is often low, especially for homology-directed repair (HDR)-mediated editing of single nucleotide polymorphisms (SNPs). Special experimental methods must be developed to circumvent this limitation; for instance, using simultaneous CRISPR/Cas9 mutagenesis and reprogramming of iPSCs (Howden et al., 2015; Tidball et al., 2018), advanced cell sorting techniques such as FACS (Forsyth et al., 2006; Miyaoka et al., 2014), or recombination of mutation-carrying cassettes though the presence of long homology arms in targeting plasmids (Hendel et al., 2014). Nevertheless, these highly specialized protocols for gene editing in iPSC lines are time and/or resource-consuming, which persists as a critical limiting factor in generating isogenic iPSC lines via CRISPR/Cas9-mediated precise SNP editing.
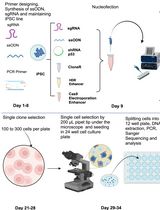
Here, we present a demonstrated workflow and detailed procedures that allow direct editing of iPSCs using the CRISPR/Cas9 system with an HDR technique by simple liposome-based transfection, followed by antibiotic selection to enrich the edited iPSCs and single clonal selection to obtain pure iPSC isogenic lines of all three possible genotypes. This method was adapted from Ran et al. (2013) with demonstrated results in our recent publications (Forrest et al., 2017; Zhang et al., 2018 and 2020). Notably, this protocol worked out very well for our unique CRISPR/cas9 SNP editing design from a heterozygous iPSC line to isogenic pairs homozygous for disease risk alleles or non-risk alleles, which makes the functional interpretation of an edited SNP more reliable by directly comparing isogenic lines of all three different genotypes (Forrest et al., 2017; Zhang et al., 2018 and 2020).
Materials and Reagents
Materials
1.5 ml Eppendorf tubes (VWR, catalog number: 89000-028)
4-w NunclonTM Delta MultiDishes (Thermo Scientific, catalog number: 62407-068)
6-well culture plates (Thermo Fisher, catalog number: 140675)
96-well culture plates (Corning, Falcon®, catalog number: 353072)
Standard 60 × 15 mm dishes w/vent (Fisher Scientific, catalog number: 12-565-95)
15 ml centrifuge tubes (VWR, catalog number: 21008-216)
2 ml cryogenic vials with closures, polypropylene (Corning®, catalog number: 89089-764)
10 μl racked tips, low-retention sterile (VWR, catalog number: 10017-062)
200 μl racked tips, low-retention sterile (VWR, catalog number: 76322-150)
1,000 μl low-retention tips (VWR, catalog number: 10017-090)
BioDot 96-well non-skirted PCR plates (Dot Scientific, catalog number: 650-PCR)
Reagents
mTeSR1 (STEMCELL, catalog number: 85850)
mFreSR Cryopreservation Medium (STEMCELL, catalog number: 05855)
ReLeSR (STEMCELL, catalog number: 05872)
Matrigel® hESC-Qualified Matrix (Corning®, catalog number: 354277)
FuGENE® HD Transfection Reagent (Promega, catalog number: E2311)
Accutase (STEMCELL, catalog number: 07920)
Primocin (Invitrogen, ant-pm-1)
Y-27632 dihydrochloride (R&D Systems, catalog number: 1254/1)
QIAprep Spin Miniprep Kit (250) (Qiagen, catalog number: 27106)
Qiaprep EndoFree Plasmid Maxi Kit (10) (Qiagen, catalog number: 12362)
BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, catalog number: 4337455)
DyeEx 2.0 kit (Qiagen, catalog number: 63204)
HotStarTaq DNA Polymerase (250 U) (Qiagen, catalog number: 203203)
PlasmidSafe Exonuclease (Lucigen, catalog number: E3101K)
QuickExtractTM DNA Extraction Solution (VWR, catalog number: 76081-768)
FastDigest BbsI (Thermo Fisher Scientific, catalog number: ER1101)
T7 ligase (NEB, catalog number: M0318S)
ATP solution, 100 mM (NEB, catalog number: N0451)
dNTP solution mix, 10 mM (NEB, catalog number: N0447S)
Opti-MEM I, 100 ml (Thermo Fisher Scientific, catalog number: 31985062)
Shrimp alkaline phosphatase (Thermo Fisher Scientific, catalog number: 783901000UN)
PCRX Enhancer System (Thermo Fisher, catalog number: 11495017)
Isopropanol (Sigma-Aldrich, catalog number: 190764)
Nuclease-free water, PCR grade (Thermo Fisher Scientific, catalog number: AM9937)
Teknova DNA/RNA Resuspension Buffer (TE buffer) (VWR, catalog number: 100216-886)
Invitrogen One Shot® Stbl3TM Chemically Competent E. coli cells (Thermo Fisher Scientific, catalog number: C737303)
1 kb DNA ladder (Promega, catalog number: G5711)
Equipment
C1000 Touch Thermo Cycler (Bio-Rad, model: 1851197)
Sorvall Legend XTR Centrifuge (Thermo Scientific, catalog number: 75217420)
Benchtop refrigerated centrifuge 5430R (Eppendorf, catalog number: 022620601)
Heracell 150i Tissue Culture Incubator (Thermo Fisher Scientific, catalog number: 51026283)
ABI 3730 DNA Analyzer (Thermo Fisher Scientific, model: 3730S)
Nalgene Mr. Frosty Freezing container (Sigma-Aldrich, catalog number: C1562)
Software
ApE – A plasmid Editor (M Wayne Davis, https://jorgensen.biology.utah.edu/wayned/ape/)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhang, H. and Zhang, S. (2021). CRISPR/Cas9-mediated Precise SNP Editing in Human iPSC Lines. Bio-protocol 11(12): e4051. DOI: 10.21769/BioProtoc.4051.
- Zhang, S., Zhang, H., Zhou, Y., Qiao, M., Zhao, S., Kozlova, A., Shi, J., Sanders, A. R., Wang, G., Luo, K., Sengupta, S., West, S., Qian, S., Streit, M., Avramopoulos, D., Cowan, C. A., Chen, M., Pang, Z. P., Gejman, P. V., He, X. and Duan, J. (2020). Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants.Science 369(6503): 561-565.
分类
干细胞 > 多能干细胞 > 细胞多能性
干细胞 > 多能干细胞 > 基于细胞的分析方法
细胞生物学 > 细胞工程 > CRISPR-cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link