- EN - English
- CN - 中文
Chromatographic Assays for the Enzymatic Degradation of Chitin
几丁质酶解的色谱分析
发布: 2021年05月05日第11卷第9期 DOI: 10.21769/BioProtoc.4014 浏览次数: 5448
评审: Andrea PuharThomas HahnAnonymous reviewer(s)
Abstract
Chitin is an insoluble linear polymer of β(1→4)-linked N-acetylglucosamine. Enzymatic cleavage of chitin chains can be achieved using hydrolytic enzymes, called chitinases, and/or oxidative enzymes, called lytic polysaccharide monooxygenases (LPMOs). These two groups of enzymes have different modes of action and yield different product types that require different analytical methods for detection and quantitation. While soluble chromogenic substrates are readily available for chitinases, proper insight into the activity of these enzymes can only be obtained by measuring activity toward their polymeric, insoluble substrate, chitin. For LPMOs, only assays using insoluble chitin are possible and relevant. Working with insoluble substrates complicates enzyme assays from substrate preparation to product analysis. Here, we describe typical set-ups for chitin degradation reactions and the chromatographic methods used for product analysis.
Graphical abstract:
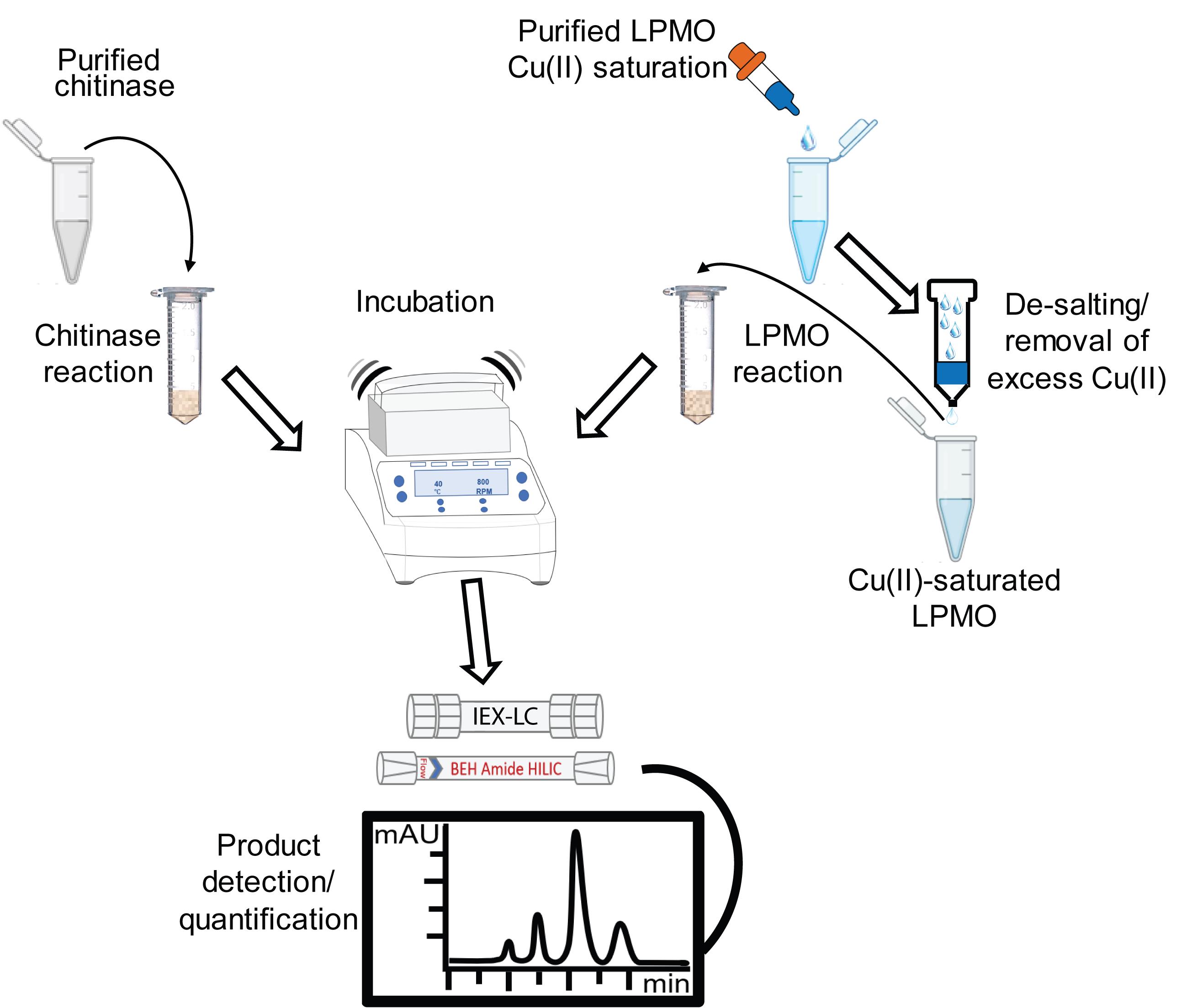
Overview of chromatographic methods for assessing the enzymatic degradation of chitin
Background
Chitin, an insoluble linear polymer of β(1→4)-linked N-acetylglucosamine, is one of the most abundant recalcitrant polysaccharides in nature, existing predominantly in two allomorphs. In α-chitin, the chains are organized in an anti-parallel fashion, which is the most common and recalcitrant form in nature; in β-chitin, the chains are organized in a parallel fashion, leading to a less recalcitrant structure with a higher water content (Gardner and Blackwell, 1975; Minke and Blackwell, 1978). Chitin is rarely found in its pure form in nature and is usually associated with minerals and proteins, and often with other polysaccharides as seen in fungal cell walls. Hence, obtaining pure chitin, e.g., from crustacean shells, requires demineralization and deproteination steps (Aye and Stevens, 2004). The efficiency of enzymatic saccharification of chitin depends on multiple factors: 1) the method used to extract chitin from the initial biomass; 2) the chitin particle size; and 3) the chitin form [e.g., Nakagawa et al. (2013)]. Thus, the choice of substrate for assaying enzymatic chitin degradation is far from trivial. It is noteworthy that the substrates used for assessing enzymatic chitin degradation, both here and in the field in general, are usually heavily processed and differ considerably from natural chitinous substrates.
Chitin may be converted to oligo- and/or mono-sugars by the coordinated action of multiple carbohydrate-active enzymes [CAZymes (Lombard et al., 2014)], which include several hydrolytic enzymes, such as chitinases (processive exo-chitinases and endo-chitinases; reviewed by Horn et al., 2006; Oyeleye et al., 2018), β-N-acetylhexosaminidases (reviewed by Slámová et al., 2010; also known as chitobiases], and oxidative metallo-enzymes called lytic polysaccharide monooxygenases [LPMOs (Vaaje-Kolstad et al., 2010)]. These enzymes differ in terms of their catalytic mechanism, substrate preferences (crystalline vs. amorphous, endo- vs. exo-attack), and product profiles (Vaaje-Kolstad et al., 2013). Chitinases cleave soluble chitooligosaccharides from chitin, the dominant product often being chitobiose. LPMOs (Chylenski et al., 2019) catalyze the oxidative cleavage of chitin chains, generating soluble and non-soluble C1-oxidized products. β-N-acetylhexosaminidases degrade soluble chitooligosaccharides produced by chitinases and LPMOs from the non-reducing end, producing N-acetylglucosamine and C1-oxidized chitobiose (chitobionic acid). In the CAZy database (Lombard et al., 2014), chitinases primarily occur in the glycoside hydrolase (GH) families GH18 and GH19. β-N-acetylhexosaminidases are found in GH family 20; however, enzymes possessing similar catalytic activity are also found in the GH families 3, 84, and 85. LPMOs are classified as Auxillary Activities (AAs), and chitin-active LPMOs have been detected in families AA10, AA11, and AA15 (Eijsink et al., 2019).
The active site of LPMOs consists of two highly conserved histidine residues that bind a single, catalytically crucial copper ion in a histidine brace (Quinlan et al., 2011; Chylenski et al., 2019). One of these histidines is the N-terminal residue (His1), which contributes to copper coordination with both its imidazole side chain and its (N-terminal) amino group. The latter implies that recombinantly produced LPMOs must be correctly processed, i.e., have an N-terminal histidine, to obtain an active enzyme (Eijsink et al., 2019). LPMOs require an oxygen-containing co-substrate (either O2 or H2O2) and a reductant for the reduction of copper (II) to copper (I). Chitin-active LPMOs appear to primarily target crystalline regions within chitin (Vaaje-Kolstad et al., 2010; Nakagawa et al., 2013), whereas chitinases are thought to prefer regions with a lower degree of substrate crystallinity. Characterization of LPMO activity faces multiple challenges related to the complex interplay among enzyme, reductant, oxidant, and substrate, as described in Eijsink et al., (2019). Importantly, LPMOs are prone to auto-catalytic inactivation (Bissaro et al., 2017; Loose et al., 2018), which obviously complicates the analysis of LPMO activity.
Proper characterization of the activity of chitinases or LPMOs, or combinations thereof, requires enzyme assays using a true polymeric substrate. While artificial chromogenic chitinase substrates are available and can be useful, measured activities toward such substrates provide only limited insight into the chitin-degrading ability of these enzymes. For chitin-active LPMOs, chromogenic artificial substrates do not exist and the very nature of these enzymes dictates that only assays using insoluble chitin are meaningful. Here, we describe methods for analyzing chitin degradation by bacterial LPMOs and chitinases. We also discuss points one should consider to successfully identify and evaluate the biochemical activities of these enzymes.
Materials and Reagents
2.0 ml microcentrifuge tubes (e.g., Axygen, catalog number: MCT-2000-C-S)
Single-channel mechanical pipettes, e.g.
0.2-2 µl (VWR, catalog number: 613-5258)
0.5-10 µl (VWR, catalog number: 613-5259)
2-10 µl (VWR, catalog number: 613-5260)
20-200 µl (VWR, catalog number: 613-5263)
100-1,000 µl (VWR, catalog number: 613-5265)
1,000-5,000 µl (VWR, catalog number: 613-5266)
Standard pipette tips, e.g.
Volume 0.1-10 µl (VWR, catalog number: 613-0735)
Volume 20-200 µl (VWR, catalog number: 613-0732)
Volume 100-1,250 µl (VWR, catalog number: 613-0739)
Volume 1,000-5,000 µl (VWR, catalog number: 613-0338)
Wide-orifice pipette tip refill system, e.g.
Volume 200 µl (VWR, catalog number: 732-3345)
Volume 1,000 µl (VWR, catalog number: 732-3348)
PD MidiTrap G-25 column (Cytiva LifeSciences, catalog number: 28918008)
Blue-capped flasks 25-1,000 ml (Fisher Scientific, DWK Life Sciences)
96-well filter plate (Millipore, catalog number: MSHVN4550) with MultiScreen Vacuum Manifold (Millipore, catalog number: MSVMHTS00)
Rezex RFQ-Fast Acid H+ (8%) 100 mm LC column (Phenomenex, catalog number: 00d-0223-k0)
Rezex RFQ-Fast Acid H+ (8%) 50 mm LC Guard column (Phenomenex, catalog number: 03b-0223-k0)
Rezex ROA-Organic Acid H+ (8%) 300 mm LC column (Phenomenex, catalog number: 00h-0138-k0)
0.3 ml polypropylene HPLC vials with caps and septa (Thermo Scientific, catalog number: 055428)
Bottle-top vacuum filtration PES filter, 0.2 or 0.45 µm (e.g., VWR, catalog number: 514-0338 or 514-0339)
Appropriate preparations of chitinases (e.g., Vaaje-Kolstad et al., 2013), chitobiase (e.g., Loose et al., 2014), and/or chitin-active LPMOs (e.g., Vaaje-Kolstad et al., 2010)
Tris (hydroxymethyl) aminomethane (Tris-HCl) (Sigma-Aldrich, catalog number: 1.08219)
Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: SX0607N)
α-Chitin from shrimp shell (e.g., from Chitinor AS or from Sigma, catalog number: C9213)
β-Chitin from squid pen (Suppliers are France Chitin, and several Japanese and Chinese companies; the preparation process is described in Chaussard and Domard (2004))
BisTris (VWR, catalog number: 0715)
Ascorbic acid (Sigma-Aldrich, catalog number: A5960)
Copper (II) sulfate (Sigma-Aldrich, catalog number: 451657)
TraceSelect water (Fisher Scientific, catalog number: 95305)
N-acetylglucosamine (GlcNAc), purity ≥99% (Sigma-Aldrich, catalog number: A8625)
Diacetyl-chitobiose (GlcNAc2), purity >95% (Megazyme, catalog number: O-CHI2)
Triacetyl-chitotriose (GlcNAc3), purity >95% (Megazyme, catalog number: O-CHI3)
Tetraacetyl-chitotetraose (GlcNAc4), purity >95% (Megazyme, catalog number: O-CHI4)
Pentaacetyl-chitopentaose (GlcNAc5), purity >95% (Megazyme, catalog number: O-CHI5)
Hexaacetyl-chitohexaose (GlcNAc6), purity >95% (Megazyme, catalog number: O-CHI6)
Chitooligosaccharide oxidase from Fusarium graminearum as described by Heuts et al. (2008)
Sulfuric acid (Sigma-Aldrich, catalog number: 258105)
Hydrochloric acid, 37% (Merck, catalog number: 100317)
Acetonitrile (VWR, catalog number: 83640.400)
Ascorbic acid stock solution (see Recipes)
20 mM Tris pH 8.0 (see Recipes)
15 mM Tris pH 8.0 (see Recipes)
1 M BisTris pH 6.0 (see Recipes)
20 ml 50 mg/ml (w/v) chitin suspension (see Recipes)
50 mM H2SO4 (see Recipes)
5 mM H2SO4 (see Recipes)
Equipment
Magnetic stirrer (e.g., IKA RCT Basic, catalog number: 0003810000)
Stirring magnets, 25 mm (e.g., VWR, catalog number: 442-4524)
Security Guard cartridge and holder (Phenomenex, catalog numbers: aj0-4490 and kj0-4282)
Acquity UPLC BEH Amide column, 130 Å, 1.7 µm, 2.1 mm × 150 mm (Waters Corp., catalog number: 186004802)
Acquity UPLC BEH Amide VanGuard pre-column, 130 Å, 1.7 µm, 2.1 mm × 5 mm (Waters Corp., catalog number: 186004799)
pH meter (e.g., Metrohm, catalog number: 2.913.0210)
4°C refrigerator
-20°C freezer
Water purification system (MilliQ water)
Benchtop centrifuge (e.g., Eppendorf centrifuge 5418/5418R, catalog number: EP022620304)
Planetary ball-mill (e.g., Retsch, PM 100, catalog number: 20.540.0001)
Grinding jar (e.g., Retsch, Zirconium Oxide 500 ml, Comfort, catalog number: 01.462.0227)
Grinding balls (e.g., Retsch, Zirconium Oxide 10 mm Ø, catalog number: 22.455.0009)
Stainless-steel sieve, 0.8 mm (e.g.,Thermo Fisher Scientific, catalog number: 10739122)
Thermomixer C (Eppendorf, catalog number: 5382000015) with ThermoTop (Eppendorf, catalog number: 5308000003) and SmartBlock 1.5/2.0 ml (Eppendorf, catalog number: 5362000035)
Vacuum pump/compressor VCP 130 (VWR, catalog number: 181-0308)
UltiMateTM 3000 UHPLC system (Thermo Fisher Scientific) with the following central parts:
SRD-3200 Solvent Rack with 2 degasser channels (Thermo Fisher Scientific, catalog number: 5035.9250)
UltiMateTM ISO-3100BM Biocompatible Isocratic Pump (Thermo Fisher Scientific, catalog number: 5042.0011)
UltiMateTM TCC-3000RS Rapid Separation Thermostatted Column Compartment (Thermo Fisher Scientific, catalog number: 5730.0000)
WPS-3000 TSL Analytical Split-Loop Thermostatted Well Plate Autosampler (Thermo Fisher Scientific, catalog number: 5822.0020)
VWD-3100 Variable Wavelength Detector, one channel (Thermo Fisher Scientific, catalog number: 5074.0005)
UHPLC Agilent Technologies 1290 Infinity (Agilent Technologies Inc., catalog number: G4220-90301)
Software
Chromeleon data system, Chromeleon 7.2.9 (Thermo Fisher Scientific, https://www.thermofisher.com/order/catalog/product/CHROMELEON7#/CHROMELEON7)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Mekasha, S., Tuveng, T. R., Vaaje-Kolstad, G. and Eijsink, V. G. H. (2021). Chromatographic Assays for the Enzymatic Degradation of Chitin. Bio-protocol 11(9): e4014. DOI: 10.21769/BioProtoc.4014.
- Mekasha, S., Tuveng, T. R., Askarian, F., Choudhary, S., Schmidt-Dannert, C., Niebisch, A. and Eijsink, V. G. H. (2020). A trimodular bacterial enzyme combining hydrolytic activity with oxidative glycosidic bond cleavage efficiently degrades chitin. J Biol Chem 295(27): 9134-9146.
分类
生物化学 > 糖类 > 多糖
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link