- EN - English
- CN - 中文
Identification of Intrinsic RNA Binding Specificity of Purified Proteins by in vitro RNA Immunoprecipitation (vitRIP)
体外RNA免疫沉淀(vitRIP)鉴定纯化蛋白的RNA结合特异性
发布: 2021年03月05日第11卷第5期 DOI: 10.21769/BioProtoc.3946 浏览次数: 4640
评审: Matilda F. ChanLifeng LiuAnonymous reviewer(s)

相关实验方案
膜蛋白–配体相互作用研究的计算流程:以含 α5 亚基的 GABA(A) 受体为例
Syarifah Maisarah Sayed Mohamad [...] Ahmad Tarmizi Che Has
2025年11月20日 2102 阅读
Abstract
RNA-protein interactions are often mediated by dedicated canonical RNA binding domains. However, interactions through non-canonical domains with unknown specificity are increasingly observed, raising the question how RNA targets are recognized. Knowledge of the intrinsic RNA binding specificity contributes to the understanding of target selectivity and function of an individual protein.
The presented in vitro RNA immunoprecipitation assay (vitRIP) uncovers intrinsic RNA binding specificities of isolated proteins using the total cellular RNA pool as a library. Total RNA extracted from cells or tissues is incubated with purified recombinant proteins, RNA-protein complexes are immunoprecipitated and bound transcripts are identified by deep sequencing or quantitative RT-PCR. Enriched RNA classes and the nucleotide frequency in these RNAs inform on the intrinsic specificity of the recombinant protein. The simple and versatile protocol can be adapted to other RNA binding proteins and total RNA libraries from any cell type or tissue.
Graphic abstract: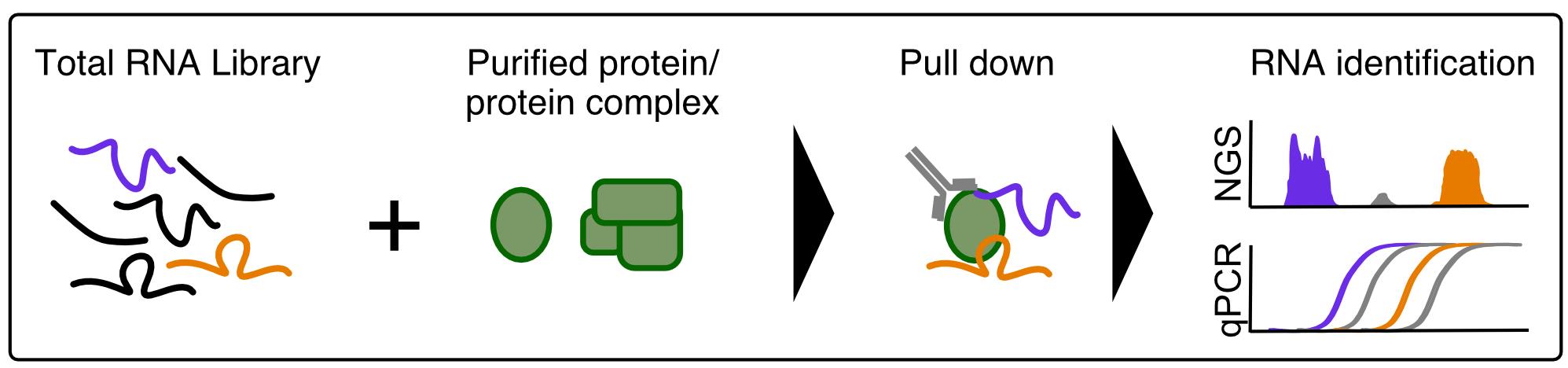
Figure 1. Schematic of the in vitro RNA immunoprecipitation (vitRIP) protocol
Background
Eukaryotic cells contain a number of different RNA classes with thousands of RNA species and a highly diverse set of proteins interacting with those. RNA-protein interactions can be classified as specific and nonspecific, depending on the definition of the bound RNA sequence or structure and on the protein domains involved in the interaction (Jankowsky and Harris, 2015). RNA interactions through non-canonical RNA binding domains with unknown specificity are increasingly observed, which raises the question how dedicated RNA targets are recognized.
A better understanding of the function of a particular RNA binding protein would thus be facilitated by the identification of its intrinsic RNA binding specificity. Since RNA binding in vivo is often modulated by cooperating factors, intrinsic specificity can only be determined in an in vitro approach (Sloan and Bohnsack, 2018). Several high-throughput techniques, such as RNAcompete (Ray et al., 2013), RNA Bind-n-Seq (Lambert et al., 2014), and in vitro iCLIP (Sutandy et al., 2018) are able to determine the RNA binding profiles of individual proteins in vitro. These methodologies use RNA oligonucleotide libraries or pools of in vitro-transcribed RNA as substrates. Such artificial RNA libraries may be highly complex, but do not represent the cellular RNA pool and may not contain secondary structures.
We developed an in vitro RNA immunoprecipitation assay (vitRIP) that uncovers intrinsic RNA-binding specificities of isolated proteins in the context of the total cellular RNA pool (Figure 1) (Müller et al., 2020). Applying vitRIP to the DExH helicase MLE (maleless) and the male-specific lethal dosage compensation complex (MSL-DCC) from Drosophila melanogaster revealed the mechanism of specific incorporation of the long non-coding roX (RNA on the X) RNAs into the MSL-DCC (Müller et al., 2020). The simple vitRIP methodology identifies transcripts bound to a recombinant protein in native conditions (no crosslinking involved) by deep sequencing or quantitative RT-PCR. vitRIP uses the complex cellular transcriptome as substrate and informs on the intrinsic binding specificity of a given protein outside of its physiological context. Furthermore, the experimental setting can be easily controlled and adapted to the research question. The following protocol details the vitRIP procedure using the RNA helicase MLE and total RNA extracted from male Drosophila S2 cells as an example (Müller et al., 2020). However, we propose that vitRIP can be applied to any RNA binding protein, which can be produced recombinantly and purified to near-homogeneity. Moreover, total RNA extracted from any cell type or tissue can serve as an RNA library.
Materials and Reagents
Note: Please ensure that all reagents and materials are RNase-free.
1.5-ml low-binding tubes (Sarstedt, catalog number: 72.706.700 )
Drosophila melanogaster S2 cells (Drosophila Genomics Resource Center, https://dgrc.bio.indiana.edu/Home) or any cell line/tissue
RNeasy Mini Kit (Qiagen, catalog number: 74104 )
Note: We did not test other RNA extraction kits, but would assume the manufacturer is not critical. Nevertheless, the extractability of various RNA species might differ between different kits.
Anti-FLAG M2 Affinity Gel (Sigma, catalog number: A2220 ) or other appropriate affinity resin, depending on the tag of the recombinant protein
Yeast tRNA solution (Sigma, catalog number: R5636 )
BSA fraction V powder (for blocking) (Sigma, catalog number: 0 5482 )
Nuclease-free water (Thermo Fisher Scientific, catalog number: AM9937 )
DNase I recombinant, RNase-free (Sigma, catalog number:0 4716728001 )
BSA 20 mg/ml solution (for vitRIP) (New England Biolabs, catalog number: B9000S )
Recombinant RNasin RNase Inhibitor (40 U/μl) (Promega, catalog number: N2515 )
Adenosine 5’-triphosphate disodium salt (ATP) (Sigma, catalog number: 10127523001 )
Proteinase K (BioCat, catalog number: BIO-37039-BL )
10% SDS solution
Phenol:chloroform:phenylalcohol ROTI Aqua-P/C/I (Carl Roth, catalog number: X985.1 )
3 M sodium acetate pH 5.2
Glycogen (20 mg/ml) (Sigma, catalog number: 10901393001 )
Ethanol absolute for analysis (Sigma, catalog number: 1009832500 )
SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific, catalog number: 18080051 )
Fast SYBR Green Mastermix (Thermo Fisher Scientific, catalog number: 4385610 )
Quant-iT Qubit RNA HS Assay kit (Thermo Fisher Scientific, catalog number: Q32855 )
Agilent RNA 6000 Pico Kit (Agilent, catalog number: 5067-1513 )
Agilent DNA 1000 Kit (Agilent, catalog number: 5067-1504 ) or Agilent High Sensitivity DNA Kit (Agilent, catalog number: 5067-4626 )
NEBNext rRNA Depletion Kit (Human/Mouse/Rat) (New England Biolabs, catalog number: E6310 )
NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs, catalog number: E7760 )
KCl
NaCl
Na2HPO4
KH2PO4
HEPES
MgCl2
NP40 (IGEPAL CA-630, Sigma, catalog number: I3021 )
Anti-FLAG M2 Monoclonal Antibody (Sigma, catalog number: F3165 )
2× SDS PAGE sample buffer (125 mM Tris HCl pH 6.8, 4% SDS, 20% glycerol, 0.01% bromophenol blue, 200 mM DTT)
1× PBS (see Recipes)
vitRIP-100 buffer (see Recipes)
vitRIP-250 buffer (see Recipes)
Equipment
-80 °C freezer
DeNovix DS-11 spectrophotometer (Biozym, catalog number: 31DS-11 ) or other spectrophotometer with 254 nm and 280 nm wavelength
Refrigerated centrifuge (Eppendorf, model: 5424R )
Eppendorf ThermoMixer C with heated lid (Eppendorf, catalog numbers: 5382000015 , 5308000003 )
Rotator (Neolab, catalog number: 2-1175 )
Qubit fluorometer (Invitrogen)
Bioanalyzer 2100 Instrument (Agilent)
Lightcycler 480 Instrument II, 384-well (Roche, catalog number: 0 5015243001 ) or another Real-Time PCR System
(Access to) Illumina HiSeq 1500 Sequencing Platform
Software
STAR – Spliced Transcript Alignment to a Reference (Alexander Dobin, Cold Spring Harbor Laboratory, https://github.com/alexdobin/STAR) (Dobin et al., 2013)
Samtools (Genome Research Limited, Sanger Institute, http://www.htslib.org) (Li et al., 2009)
Bedtools (Quinlan laboratory, University of Utah, https://bedtools.readthedocs.io/en/latest/)
IGV – Integrative Genomics Viewer and igvtools (IGV Team, Broad Institute and UC San Diego, http://software.broadinstitute.org/software/igv/home) (Robinson et al., 2011)
R (R Core Team, https://www.r-project.org)
RStudio (RStudio, https://rstudio.com)
Bioconductor (Bioconductor Core Team, http://bioconductor.org) (Huber et al., 2015)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Müller, M., Schauer, T. and Becker, P. B. (2021). Identification of Intrinsic RNA Binding Specificity of Purified Proteins by in vitro RNA Immunoprecipitation (vitRIP). Bio-protocol 11(5): e3946. DOI: 10.21769/BioProtoc.3946.
分类
生物化学 > 蛋白质 > 相互作用 > Protein-RNA 相互作用
生物化学 > RNA > RNA-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link