- EN - English
- CN - 中文
Rapid Genome Engineering of Pseudomonas Assisted by Fluorescent Markers and Tractable Curing of Plasmids
受荧光标记和易于固化质粒影响的假单胞菌快速基因组工程
发布: 2021年02月20日第11卷第4期 DOI: 10.21769/BioProtoc.3917 浏览次数: 7377
评审: Miaowei Marwell MaoKhyati Hitesh ShahAnonymous reviewer(s)
Abstract
Precise genome engineering has become a commonplace technique for metabolic engineering. Also, insertion, deletion and alteration of genes and other functional DNA sequences are essential for understanding and engineering cells. Several techniques have been developed to this end (e.g., CRISPR/Cas-assisted methods, homologous recombination, or λ Red recombineering), yet most of them rely on the use of auxiliary plasmids, which have to be cured after the editing procedure. Temperature-sensitive replicons, counter-selectable markers or repeated passaging of plasmid-bearing cells have been traditionally employed to circumvent this hurdle. While these protocols work reasonably well in some bacteria, they are not applicable for other species or are time consuming and laborious. Here, we present a fast and versatile protocol of fluorescent marker-assisted genome editing in Pseudomonas putida, followed by clean curing of auxiliary plasmids through user-controlled plasmid replication. One fluorescent marker facilitates identification of genome-edited colonies, while the second reporter enables detection of plasmid-free bacterial clones. Not only is this protocol the fastest available for Pseudomonas species, but it can be easily adapted to any type of genome modifications, including sequence deletions, insertions, and replacements.
Graphical abstract:
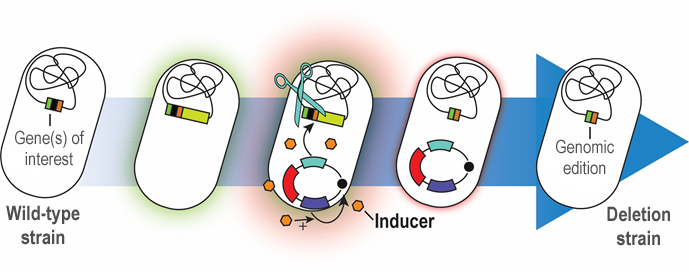
Rapid genome engineering of Pseudomonas with curable plasmids
Background
Targeted, precise genomic manipulation techniques have considerably advanced the field of microbial engineering. Such methods not only allow for assessing genotype-phenotype relationships, but also enable complex engineering of microbial cell factories. In recent years, CRISPR/Cas9 approaches have paved the way for precise genome engineering in eukaryotes. In bacteria, the use of CRISPR/Cas9 is mainly limited to its value as a counter-selection tool, as bacteria lack non-homologous end-joining to repair the double-strand breaks induced by the Cas9 nuclease. Therefore, engineering efforts in many bacteria rely on homologous recombination (HR) to alter the genome. The advantage of HR is that a broad range of alterations can be introduced in the target genome. Furthermore, it is applicable not only to the so-called model organisms, e.g., Escherichia coli and Saccharomyces cerevisiae, but also finds wide spread application in non-traditional hosts, e.g., Pseudomonas species. In this protocol, we provide a workflow for HR-based genome engineering of P. putida – paired with an advanced toolbox that includes several resistance markers – facilitated by the use of fluorescent markers that enable monitoring of every step (Wirth et al., 2020). The presented methodology relies on the co-integration of a suicide plasmid [controlled by the pir-dependent origin of replication ori(R6K)] at the locus of interest. The co-integration locus is determined by two homologous arms (HAs) on the suicide plasmid, which can be freely chosen by the user to mediate HR. A resolving step forces a second HR event that leads to removal of the plasmid backbone from the genome. This step is triggered by the action of the homing endonuclease I-SceI, acting on two recognition sequences that flank the homologous regions within the backbone of the suicide plasmid. The gene encoding I-SceI is supplied in trans from a helper plasmid, introduced into the cells after co-integration of the suicide plasmid. Our recently developed method facilitates rapid curing of this auxiliary plasmid through a synthetic, controllable replication mechanism (Volke et al., 2020) dependent on the presence of 3-methylbenzoic acid (3-mBz). Therefore, plasmid replication can be tightly regulated by the user by merely supplementing or omitting the inducer molecule in the culture medium. Plasmid curing is further aided by the expression of a fluorescent marker from the auxiliary vector, which is compatible with the reporter gene employed in the suicide plasmid. To broaden the use of this method, we developed different versions of the involved plasmids with several antibiotic resistance markers.
Materials and Reagents
Material
Pipette tips (1,000 μl, 200 μl, 10 μl) (Sartorius, catalog numbers: 7902020 , 790012 , 791002 )
Sterile Petri dishes (Ø = 90 mm) (HiMedia Laboratories, catalog number: PW001 )
Eppendorf tubes (Tarsons Products, 1.5 ml, 2.0 ml)
50-ml conical tubes (Sarstedt, catalog number: 62.547.205 )
Electrocuvettes, 0.1-cm gap for E. coli (Bio-Rad, Gene Pulser, catalog number: 165-2089) and 0.2-cm gap for Pseudomonas (Bio-Rad, Gene Pulser, catalog number: 165-2086 )
Sterile 0.2-μm syringe filters (Sigma-Aldrich)
3-Methylbenzoic acid (synonym m-toluic acid) (3-mBz; Sigma-Aldrich, ReagentPlus, catalog number: T36609 )
Sucrose (Sigma-Aldrich, Milipore, catalog number: 84100 )
Lysogeny broth (LB) (Sigma-Aldrich, catalog number: L3522 ); preparation according to the manufacturer’s instructions, storage for up to three weeks at room temperature
Agarised LB (Sigma-Aldrich, catalog number: L3147 ); preparation according to the manufacturer’s instruction, storage for up to two months at 4 °C
SOC medium (Sigma-Aldrich, catalog number: S1797 )
Kanamycin (TH-Geyer, catalog number: T832.3 )
Gentamicin (Sigma-Aldrich, catalog number: G1264 )
Streptomycin (Sigma-Aldrich, catalog number: S6501 )
Ampicillin (Mitolab, catalog number: K029 )
Oligonucleotides (Integrated DNA Technologies, Leuven, Belgium)
Uracil-specific excision reagent (NEB Biolabs, USER enzyme, catalog number: M5505 )
DNA polymerase (Thermo Fisher Scientific, Phusion U Hot start, catalog number: F555 )
DNA polymerase reaction mix for colony PCRs including (NEB Biolabs, OneTaq® Hot Start Quick-Load® 2× master mix with standard buffer, catalog number: M0488L )
Reagents to prepare chemically competent E. coli cells (Zymoresearch, Mix & Go!; catalog number: T3001 )
Escherichia coli DH5α λpir [endA1 hsdR17 glnV44 (supE44) thi-1 recA1 gyrA96 relA1 ϕ80dlacΔ(lacZ)M15 Δ(lacZYA-argF)U169 zdg-232::Tn10 uidA::pir+] (Platt et al., 2000)
P. putida KT2440 (strain ATCC 47054/DSM 6125/NCIMB 11950) (Belda et al., 2016)
Sequencing kit (Eurofins, Mix2Seq Kit OVERNIGHT, catalog number: 3094-0ONMSK )
Plasmid purification kit (Macherey-Nagel, NucleoSpin Plasmid, catalog number: 740588 )
Gel and PCR Clean-up Kit (Macherey-Nagel, NucleoSpin™ Gel and PCR Clean-up Kit, catalog number: 740588 )
Optional: DpnI (Thermo Fisher Scientific, FastDigest DpnI, catalog number: FD1703 )
Agarose (Bio-Rad, Certified Molecular Biology Agarose, catalog number: 1613102); prepare at 1% (w/v) in 1× TAE buffer for gel electrophoresis (use microwave heating to dissolve ). Can be stored at 60 °C to keep molten for immediate use
Fluorescent nucleic acid staining solution (Intronbio, Red safe, catalog number: 21141 )
DNA ladder (Thermo Fisher Scientific, GeneRuler 1 kB, catalog number: SM0314 )
3-Methylbenzoic acid (3-mBz) solution (500 mM) (see Recipes)
Sucrose solution (300 mM) (see Recipes)
Polymerase chain reaction (PCR) reagents (see Recipes)
Electro-competent Pseudomonas cells (see Recipes)
Antibiotic stock solutions (see Recipes)
Equipment
Electroporator (Bio-Rad, MicroPulser, catalog number: 1652100 )
Transilluminator (Thermo Fisher Scientific, Safe Imager 2.0 Blue Light, catalog number: G6600 )
Table centrifuge, used for 1.5- and 2-ml reaction tubes (VWR, model: Microstar 17R, catalog number: 521-1647 )
Table centrifuge, used for 50-ml reaction tubes (Thermo Fisher Scientific, model: Heraeus Multifuge X1R, catalog number: 75004250 ) with rotor (Thermo Fisher Scientific, model: TX-400, catalog number: 75003181 ) and buckets (Thermo Fisher Scientific, catalog number: 75003655 ) and adaptors (Thermo Fisher Scientific, catalog number: 75003683 )
Termoblock (Eppendorf, model: Thermo Mixer C, catalog number: 5382000015 )
pH-meter (Thermo Fisher Scientific, model: FE150pH, catalog number: S35924 )
Agarose chamber, gelcaster, combs and power supply (Bio-Rad, Mini-sub Cell GT system & Power pack, catalog number: 1645050 )
Gel visualization (Bio-Rad, Gel Doc XR+ Gel Documentation System, catalog number: 1708195EDU )
PCR thermocycler (Eppendorf, Mastercycler Nexus X2 Thermocycler, catalog number: 6336000015 )
Software
AMUSER [http://www.cbs.dtu.dk/services/AMUSER (Genee et al., 2015)]
DNA sequence design tool for example Benchling or Geneious (Biomatter Ltd.)
Procedure
文章信息
版权信息
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Volke, D. C., Wirth, N. T. and Nikel, P. I. (2021). Rapid Genome Engineering of Pseudomonas Assisted by Fluorescent Markers and Tractable Curing of Plasmids. Bio-protocol 11(4): e3917. DOI: 10.21769/BioProtoc.3917.
分类
微生物学 > 微生物生物化学 > DNA
分子生物学 > DNA > 诱/突变
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link