- EN - English
- CN - 中文
Equilibrium and Kinetic Measurements of Ligand Binding to HiBiT-tagged GPCRs on the Surface of Living Cells
配体与HiBiT标记的GPCR在活细胞表面上的平衡和动力学测量
发布: 2020年12月20日第10卷第24期 DOI: 10.21769/BioProtoc.3861 浏览次数: 4791
评审: David A. CisnerosAndrea GramaticaAlberto Rissone
Abstract
G-protein coupled receptors (GPCRs) remain at the forefront of drug discovery efforts. Detailed assessment of features contributing to GPCR ligand engagement in a physiologically relevant environment is imperative to the development of new therapeutics with improved efficacy. Traditionally, binding properties such as affinity and kinetics were obtained using biochemical radioligand binding assays. More recently, the high specificity of resonance energy transfer has been leveraged toward the development of homogeneous cell-based proximity assays with capacity for real-time kinetic measurements. This suite of ligand binding protocols couples the specificity of bioluminescent resonance energy transfer (BRET) with the sensitivity afforded by the luminescent HiBiT peptide. The BRET format is used to quantify dynamic interactions between ligands and their cognate HiBiT-tagged GPCRs through competitive binding with fluorescent Tracers. At the same time, high affinity complementation of HiBiT with the cell impermeable LgBiT limits the bright bioluminescence donor signal to the cell surface and eliminates luminescence background from unoccupied receptors present in intracellular compartments.
Keywords: HiBiT (HiBiT)Background
G protein-coupled receptors (GPCRs) play critical roles in driving cell signaling pathways and remain a prominent target class for drug development (Santos et al., 2017). Accordingly, GPCR ligand binding assays that can quantify biophysical properties critical to drug action are essential for both basic research and drug discovery campaigns (Fang, 2012; Stoddart et al., 2016). Biochemical assessment of ligand binding affinity under equilibrium conditions has traditionally been viewed as an acceptable indicator for in vivo pharmacology of drug candidates (Hulme and Trevethick, 2010). However, there is growing understanding that this approach might be too simplistic. The kinetics of ligand binding provides insights into duration of drug action including residence time and potential for rapid rebinding, which have been postulated to be relevant predictors for in vivo efficacy (Fang, 2012; Copeland, 2016; Sykes et al., 2019). In addition, many GPCRs function as parts of multi-protein signaling complexes, which might influence distinct characteristics of ligand binding (Kenakin, 2010; Fang, 2012). As a result, there is increasing interest in comprehensive measurements of GPCR ligand engagement in the physiological context of living cells in both equilibrium and real-time.
GPCR ligand binding assays routinely employ ligands labeled with either radioisotopes or fluorophores in order to track the binding to a target (Fang, 2012; Stoddart et al., 2016). The most common biochemical GPCR ligand binding assays utilize radioligands in a competitive binding format to measure interactions between ligands and their unmodified cognate GPCRs (Flanagan, 2016). The specificity of these assays relies on the use of highly selective radioligands in combination with overexpressed GPCRs within membrane preparations. In addition, these non-homogenous assays require separation of free from bound radioligand, which limits the resolution and throughput of kinetic analyses (Sykes et al., 2019). Furthermore, the wash steps are more difficult to perform in high throughput screenings.
Proximity binding assays relying on time resolved fluorescence resonance energy transfer (TR-FRET) or bioluminescent resonance energy transfer (BRET) have emerged as attractive alternatives to radioligand binding assays, particularly for high throughput screenings (Stoddart et al., 2016; Sykes et al., 2019). These assays typically use fluorescently labeled ligands (fluorescent Tracers) in a competitive binding format to measure interactions between unmodified ligands and their cognate GPCRs genetically fused to an energy donor (i.e., a lanthanide-labeled protein tag or a luciferase). Although both ligand and GPCR need to be modified, these approaches provide high specificity as signal is generated only when the fluorescent Tracer and tagged GPCR are in close proximity. The inherent distance constraints of energy transfer (Ciruela, 2008) eliminates the requirement for highly selective labeled ligands and permits homogeneous cell-based assays. The homogeneous format allows for real-time kinetic measurements and is generally more compatible with high throughput settings.
The development of the bright NanoLuc luciferase (Hall et al., 2012) enabled BRET-based GPCR ligand binding assays previously impossible with other luciferases (Stoddart et al., 2015; Soave et al., 2016). Still, due to the cell permeability of the NanoLuc substrate furimazine, the use of full length NanoLuc as the energy donor results in signal generation regardless of NanoLuc’s cellular localization. This could lead to an elevated background from NanoLuc-tagged receptors that are present in intracellular compartments and are unable to engage the Tracer. This issue was recently addressed by genetically fusing GPCRs to the luminescent peptide HiBiT rather than the full length NanoLuc (Soave et al., 2019; Boursier et al., 2020). HiBiT is a small 11-amino acid peptide that produces bright luminescence upon high affinity complementation with LgBiT, an 18 kDa subunit derived from NanoLuc (Schwinn et al., 2018). The use of HiBiT restricts signal generation in endpoint assays to the cell surface. This is because LgBiT is cell impermeable and therefore incapable to complement any HiBiT-tagged receptors located in intracellular compartments. An added benefit is the use of a small peptide tag, which is less likely to interfere with protein function and trafficking to the cell surface.
The binding protocols described here couple the specificity of BRET with the sensitivity and inherent cell-surface localization of the HiBiT/LgBiT reporter for quantification of binding interactions with selective GPCRs on the surface of living cells (Figure 1). They include four principal types of assays using β2-AR ligand binding as an example. Two of these assays utilize increasing concentrations of a fluorescent Tracer to measure its binding characteristics in equilibrium and real-time. The other two assays employ increasing concentrations of an unmodified test compound to derive its binding properties through the competition with a fixed concentration of a fluorescent Tracer. These competition analyses can be performed under equilibrium or in a kinetic format, which take advantage of a method reported by Motulsky and Mahan (1984).
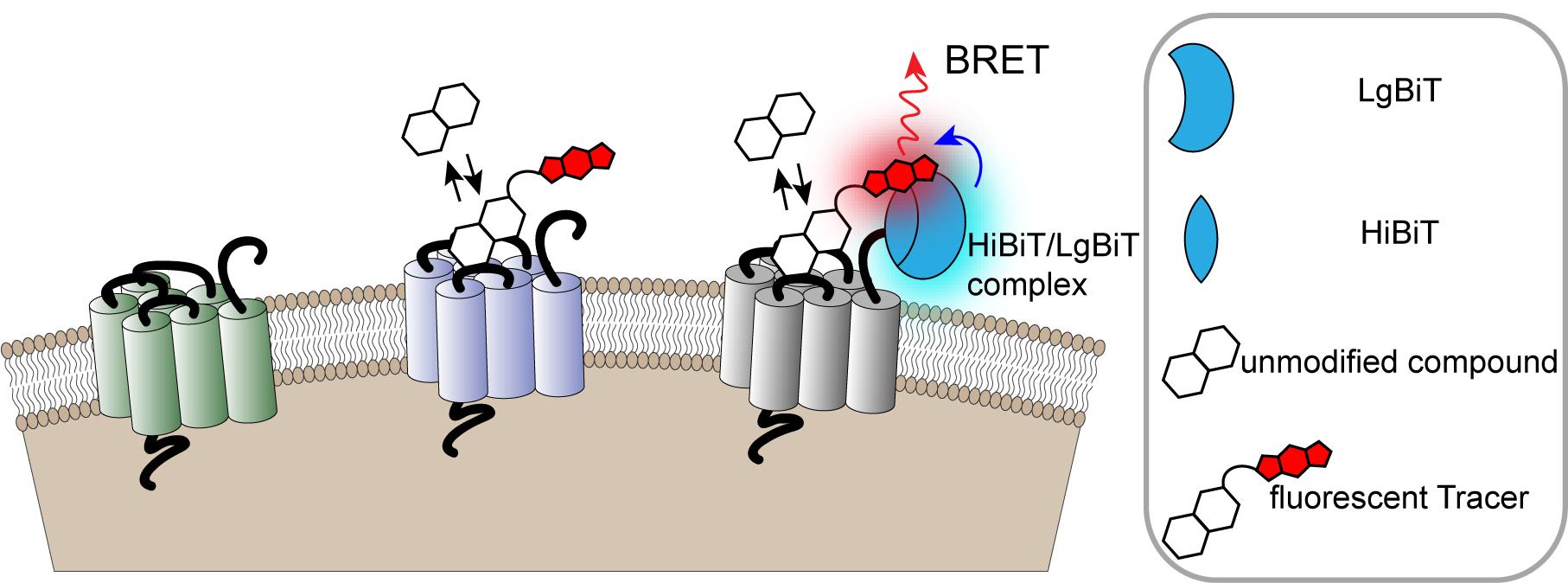
Figure 1. Monitoring cell surface ligand engagement with selective HiBiT-tagged GPCRs via BRET. BRET assay utilizing fluorescent Tracers in a competitive binding format to quantify dynamic cell-surface interactions between ligands and their cognate HiBiT-tagged GPCRs. This approach benefits from high specificity as signal is generated only when the fluorescent Tracer and tagged GPCR are in close proximity.
Materials and Reagents
White, Tissue Culture-Treated (TC) 96-well plates (Corning, catalog number: 3917 )
Non-binding V bottom plates (for serial dilutions; for example, Costar, catalog number: 3896 )
Tissue Culture-Treated (TC) flasks (growth area 150 or 75 cm2; Falcon, catalog number: 355001 or 353107 )
HEK293 cells
0.05% Trypsin/EDTA (Life Technologies, catalog number: 25300 )
Dulbecco's Modified Eagle Medium (DMEM) (Life Technologies, catalog number: 11995 )
Fetal Bovine Serum (FBS) (HyClone, catalog number: SH30070.03 )
100x Penicillin-streptomycin (Sigma, catalog number: P4333 )
Opti-MEM without Phenol Red (Life Technologies, catalog number: 11058 )
FuGENE HD (Promega, catalog number: E2311) or ViaFect (Promega, catalog number: E4981 )
Note: Other transfection reagents can also be used.DMSO (Sigma, catalog number: D2650-5X5ML )
Tracer dilution buffer (Promega, catalog number: N2191 )
Nano-Glo HiBiT Extracelular Detection System (Promega, catalog number: N2420 or N2421 )
Transfection Carrier DNA (Promega, catalog number: E4881 )
Test compound of interest
DNA construct encoding a HiBiT- tagged GPCR fusion
Note: HiBiT-tagged GPCR constructs can be obtained from Promega Custom Assays Systems or generated via cloning into pFN39K secHiBiT CMV-neo Flexi Vector (Promega, catalog number: N2411 ) or pBiT3.1-secN [CMV/HiBiT/Blast] vector (Promega, catalog number: N2381).
Fluorescent Tracer
Note: Several NanoBRET GPCR Tracers can be obtained from Promega Custom Assays Systems. In addition a fluorescent GPCR Tracer can be generated according to the published protocol (Robers et al., 2019) using building blocks that can be obtained from Promega Custom Assays Systems.
Miscellaneous tissue culture reagents
DMEM Cell Culture Medium (see Recipes)
Opti-MEM Assay Medium (see Recipes)
Equipment
BRET-compatible microplate luminometer equipped with 450 nm (bandpass) and 600 nm (longpass) filters (e.g., Promega GloMax Discover, PerkinElmer EnVision, or BMG Clariostar)
Orbital plate shaker
Miscellaneous tissue culture equipment
Software
GraphPad Prism (https://www.graphpad.com/scientific-software/prism)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Boursier, M. E., Levin, S., Hurst, R. and Friedman Ohana, R. (2020). Equilibrium and Kinetic Measurements of Ligand Binding to HiBiT-tagged GPCRs on the Surface of Living Cells. Bio-protocol 10(24): e3861. DOI: 10.21769/BioProtoc.3861.
- Boursier, M. E., Levin, S., Zimmerman, K., Machleidt, T., Hurst, R., Butler, B. L., Eggers, C. T., Kirkland, T. A., Wood, K. V. and Friedman Ohana, R. (2020). The luminescent HiBiT peptide enables selective quantitation of G protein-coupled receptor ligand engagement and internalization in living cells. J Biol Chem 295(15): 5124-5135.
分类
癌症生物学 > 通用技术 > 药物发现和分析
细胞生物学 > 基于细胞的分析方法 > 蛋白互作
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link