- EN - English
- CN - 中文
Serial Cryomicrotomy of Saccharomyces cerevisiae for Serial Electron Cryotomography
用于连续电子低温断层扫描的酿酒酵母连续冷冻切片
发布: 2020年11月20日第10卷第22期 DOI: 10.21769/BioProtoc.3831 浏览次数: 4447
评审: Ashraf Al-AmoudiKrishna SaharanAnonymous reviewer(s)
Abstract
Electron cryotomography (cryo-ET) is an increasingly popular technique to study cellular structures and macromolecules in situ. Due to poor penetration of electrons through thick biological samples, the vitreously frozen samples for cryo-ET need to be thin. For frozen-hydrated cells, such samples can be produced either by cryomicrotomy or cryo-FIB-milling. As a result, a tomogram of such a sample contains information of a small fraction of the entire cell volume, making it challenging to image rare structures in the cell or to determine the distribution of scattered structures. Here, we describe the tools and workflow that we designed to facilitate serial cryomicrotomy, which makes possible the exploration of a larger volume of individual cells at molecular resolution. We successfully used serial cryomicrotomy to locate and image the Dam1/DASH complex located at microtubule plus ends inside mitotic Saccharomyces cerevisiae cells.
Keywords: Serial electron cryotomography (连续电子低温断层扫描)Background
Chromosomes have to be coupled to kinetochore microtubules for successful segregation during cell division. The kinetochore is the macromolecular complex responsible for coupling the chromosomes to the kinetochore microtubules’ dynamic plus ends. In the budding yeast Saccharomyces cerevisiae, the Dam1/DASH complex is a part of the kinetochore that interfaces with kinetochore microtubules. There have been many studies to reconstitute kinetochore subcomplexes in vitro to understand its structure and function (Musacchio and Desai, 2017; Hinshaw and Harrison, 2018). These attempts have led to multiple models that suggest how the kinetochore functions. To test some of these outer-kinetochore models in S. cerevisiae, we use electron cryotomography (cryo-ET) to visualize the S. cerevisiae outer kinetochore at molecular resolution (4-6 nm) in situ.
Cryo-ET is an increasingly popular technique used to study cellular structures and macromolecules in situ at molecular resolution (Ng and Gan, 2020). Many of the earlier cryo-ET studies involve using biological samples that are thick (> 500 nm), resulting in reconstructions with low contrast. To increase the contrast of the reconstructions while preserving cellular context of the samples, the samples have to be much thinner, but not too thin. Therefore, a reasonable compromise is ~100 nm thickness. The two main techniques used to obtain thin samples for cryo-ET are cryo-FIB-milling and cryomicrotomy. Cryo-FIB-milling involves using gallium ions to sputter away biological material until a thin plank-like cryo-lamella of biological sample remains (Marko et al., 2006; Hayles et al., 2007; Rigort et al., 2010; Villa et al., 2013; Mahamid et al., 2015; Medeiros et al., 2018). Cryomicrotomy involves cutting a frozen-hydrated sample, such as a cell paste, into thin cryosections using a diamond knife (Al-Amoudi et al., 2004a and 2004b). Each cryo-lamella or cryosection contains only a small fraction of a cell volume. It is worth noting, however, that serial cryomicrotomy would allow us to explore a larger fraction of a cell at molecular resolution compared to cryo-FIB-milling. This can be done by imaging multiple consecutive cryosections of the same cell. Here, we describe both the tools and workflow that we designed to facilitate serial cryomicrotomy and data collection. We successfully used serial cryomicrotomy and serial cryo-ET to locate and image the Dam1/DASH complex in S. cerevisiae. We also explore the cryomicrotomy parameters that minimize artefacts in yeast cryosections. This approach has also been used to study mitotic fission yeast and meiotic budding yeast (Cai et al., 2018; Ma et al., 2019).
Materials and Reagents
Parafilm
50 ml Falcon tube
1 ml pipette tip
S. cerevisiae cells
0.1 mg/ml bovine serum albumin (Sigma-Aldrich)
10-nm gold nanoparticles (BBI Solutions, EM.GC10 )
50% (v/v) ethanol in distilled H2O, 0.2 µm filtered (500 ml)
18 MΩ H2O, 0.2 µm filtered (1 L)
Ensure that there are no debris in both H2O and ethanol before using as debris can damage the diamond knife.
Dextran (Mr ~6000) (Sigma Aldrich, catalog number: 31388 )
Pressurized ethane gas
Liquid nitrogen
Equipment
Tools (Standard)
Parallel-bar EM grids coated with continuous carbon film (Electron Microscopy Sciences, catalog number: G150PB-CU )
EM grid boxes
K100X Glow Discharge (Quorum Emitech)
Note: Product has been discontinued in 2010. The current equivalent is the SC7620 Mini Sputter Coater/Glow Discharge System.
Fine-tipped and self-locking tweezers for handling EM grids (Ted Pella, Inc.)
Metal chuck (included with UC7/FC7)
Chuck adaptor (0.45 mm) (included with UC7/FC7)
Plastic FC7 cover (included with UC7/FC7)
UC7/FC7 (Leica Microsystems)
Note: An alternative cryomicrotome by RMC Products is commercially available. However, we do not have any experience with that product.
Diamond trimming and cutting knives (Diatome)
Crion (Leica Microsystems)
Centrifuge
Diamond knife
Forceps
Tools (Non-standard)
Fibre tool (Figure 1A)
Guinea pig hair (available from pet shops or animal husbandry facilities)
Micromanipulator (Narishige Scientific Instrument Lab, catalog number: MN-151S [6 mm joystick travel]) (Figure 1B) (Inspired by Ladinsky et al., 2006 ; Ladinsky, 2010; Studer et al., 2014 )
Note: Any commercially available micromanipulator can be adapted for use in cryomicrotomy.
Copper tubes (0.45 mm inner diameter, 0.65 mm outer diameter) and wire tool for drawing samples into copper tubes (Part 733-1, Engineering Office M Wohlwend GmbH) (Figure 1C)
Flat-nosed pliers (Figure 1D)
Copper tube cutting device (Part. 732, Engineering Office M Wohlwend GmbH) (Figure 1E)
Steel bucket for storing copper tubes (Figure 1F) (design based on Zuber lab’s cryosectioning tools)
Aluminium mount (base and insert pieces) (Figure 1G)
Slotted plastic FC7 covers [modified from (Studer et al., 2014) to minimize ice contamination] (Figure 1H)
Air dust blower
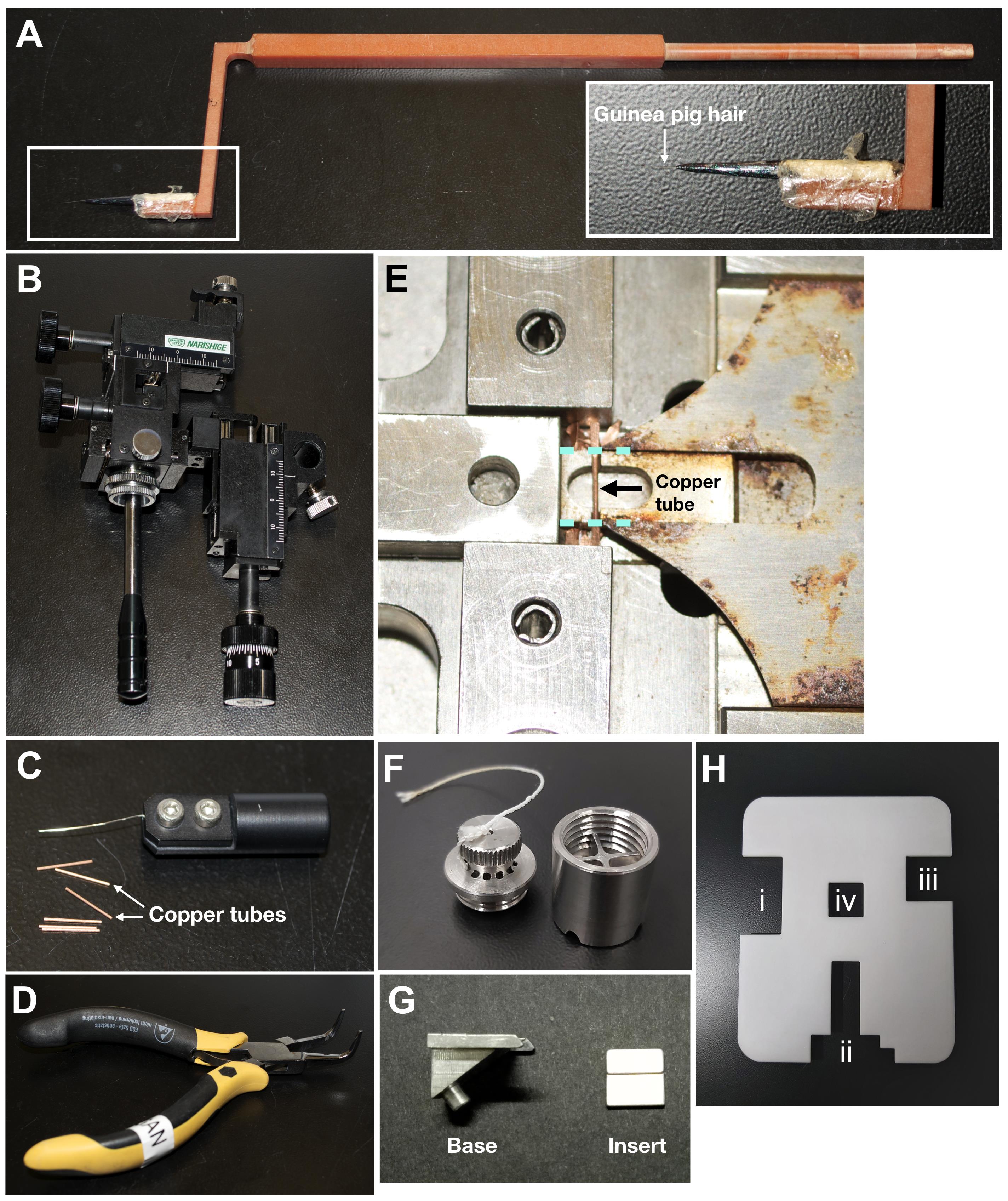
Figure 1. Tools for enabling serial cryomicrotomy. A. Custom-made fibre tool for manipulating the cryoribbon during cryomicrotomy. B. Custom micromanipulator for fine control of the fibre tool. During cryomicrotomy, the micromanipulator is secured to a heavy steel block with a magnetic stand. C. Copper tubes (0.45 mm inner diameter) and wire tool (upper) for drawing samples into the copper tubes for self-pressurized freezing. D. Pliers for clamping shut the end of the copper tubes before freezing. E. Device for cutting off flat ends of copper tube along the cyan dotted lines. Note that the rust, which cannot be completely removed, does not affect the performance of the cutting device. Users can also consider a cutter made from a rust-prone material. F. Custom steel bucket for storing copper tubes containing frozen samples. Steel bucket can easily fit into a 50 ml Falcon tube. G. Base and insert pieces of the aluminium mount that can be assembled on a diamond knife. H. Slotted plastic FC7 cover with cutouts for the (i) forceps, (ii) Crion, (iii) fibre tool, and (iv) for observing during cryomicrotomy. The translucency of the cover does not affect cryomicrotomy workflow.
Software
IMOD (Mastronarde, 1997) (https://bio3d.colorado.edu/imod/)
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ng, C. T., Ladinsky, M. and Gan, L. (2020). Serial Cryomicrotomy of Saccharomyces cerevisiae for Serial Electron Cryotomography. Bio-protocol 10(22): e3831. DOI: 10.21769/BioProtoc.3831.
- Ng, C. T., Deng, L., Chen, C., Lim, H. H., Shi, J., Surana, U. and Gan, L. (2019). Electron cryotomography analysis of Dam1C/DASH at the kinetochore-spindle interface in situ. J Cell Biol 218(2): 455-473.
分类
生物物理学 > 电子冷冻断层扫描
细胞生物学 > 细胞成像 > 冷冻超薄切片
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link