- EN - English
- CN - 中文
Fluorescent Polysome Profiling in Caenorhabditis elegans
秀丽隐杆线虫荧光多聚体分析
发布: 2020年09月05日第10卷第17期 DOI: 10.21769/BioProtoc.3742 浏览次数: 7468
评审: Madhuja SamaddarAyush RanawadeAnonymous reviewer(s)
Abstract
An important but often overlooked aspect of gene regulation occurs at the level of protein translation. Many genes are regulated not only by transcription but by their propensity to be recruited to actively translating ribosomes (polysomes). Polysome profiling allows for the separation of unbound 40S and 60S subunits, 80S monosomes, and actively translating mRNA bound by two or more ribosomes. Thus, this technique allows for actively translated mRNA to be isolated. Transcript abundance can then be compared between actively translated mRNA and all mRNA present in a sample to identify instances of post-transcriptional regulation. Additionally, polysome profiling can be used as a readout of global translation rates by quantifying the proportion of actively translating ribosomes within a sample. Previously established protocols for polysome profiling rely on the absorbance of RNA to visualize the presence of polysomes within the fractions. However, with the advent of flow cells capable of detecting fluorescence, the association of fluorescently tagged proteins with polysomes can be detected and quantified in addition to the absorbance of RNA. This protocol provides detailed instructions on how to perform fluorescent polysome profiling in C. elegans to collect actively translated mRNA, to quantify changes in global translation, and to detect ribosomal binding partners.
Keywords: C. elegans (秀丽线虫)Background
Multiple actively translating ribosomes bound to the same mRNA transcript are called polysomes. Polysomes can be separated from the other ribosomal forms and unbound mRNA using polysome profiling. Polysome profiling has been a key technique in the field of protein translation. In conjunction with mRNA-seq, polysome profiling allows for transcripts that are regulated by post-transcriptional mechanisms to be detected and quantified (Lan et al., 2019; Rollins et al., 2019) by quantitative PCR (Panda et al., 2017). Polysome profiling has also been used as a readout for global translation rates by quantifying RNA associated with polysomes under different conditions (Merret et al., 2015; Chassé et al., 2017). Additionally, polysome profiling has been used to isolate proteins that comprise polysomes and have allowed their contents to be probed using techniques like western blotting (Tiedje et al., 2012; Jin and Xiao, 2018).
This polysome profiling protocol is different from previously published protocols in two regards; it is developed specifically for C. elegans and it uses fluorescence to detect association of a fluorescently tagged protein with polysomes. Compared to cell culture or harvested tissue, the cuticle of C. elegans requires additional considerations when creating the lysate for polysome profiling.
Existing polysome profiling methods allow for associations between a protein of interest and active ribosomes to be detected. However, this requires the protein to be extracted from the polysome fractions and probed for the protein of interest using western blotting. To resolve differences in associations between a protein of interest and the different forms of ribosomes (40S, 60S, monosomes, polysomes) multiple fractions need to be collected and probed from each sample (Figure 1). Instead, the use of a flow cell capable of detecting fluorescence allows for the presence of a fluorescently tagged protein to be detected and quantified among the entire profile. Thus, associations between the tagged protein and all forms of the ribosome are captured with high resolution and the need for western blotting is eliminated. The following polysome profiling protocol will cover how to prepare, load, centrifuge, and scan sucrose gradients using lysate from C. elegans (Figure 2). Additionally, instructions are given to quantify the proportion of actively translating ribosomes in a polysome profile.
Figure 1. Polysome profiling overview. A. Ultra-centrifugation of a cell, tissue, or whole animal lysate allows separation of free ribonucleoprotein complexes (RNPs) from RNA bound to 40S (blue), 60S (red), and 80S monosomes (yellow), and polysomes (green). B. RNA absorbance measurements and fractionation are used to isolate free mRNA and mRNA bound to monosomes and polysomes.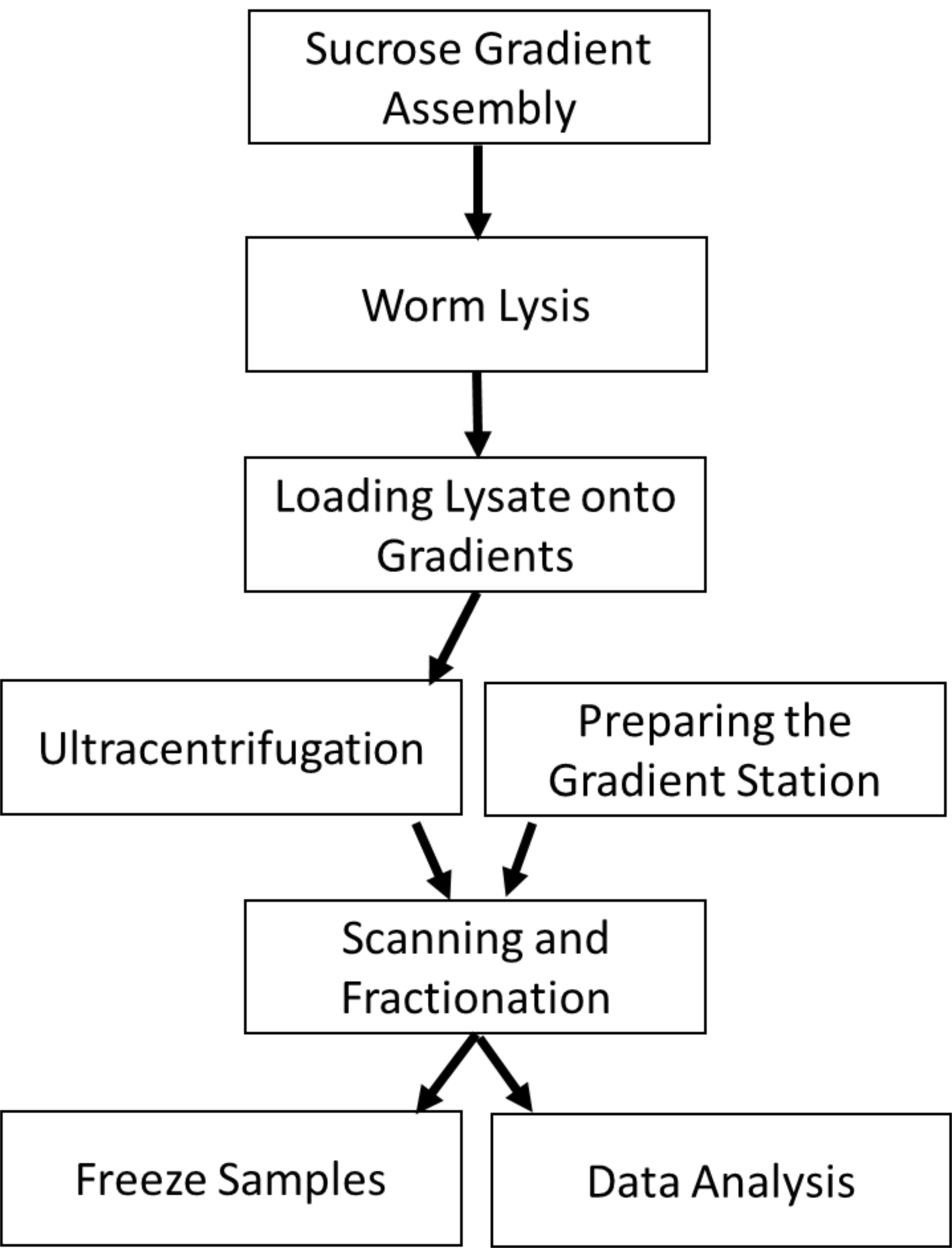
Figure 2. Protocol Overview. The protocol begins with preparing sucrose gradients onto which worm lysate will be loaded. Ultracentrifugation of the lysate allows the polysomal RNA to be sedimented away from RNA not bound by polysomes. During the centrifugation, the gradient station can be prepared by flushing the lines and blanking the flow cell. The centrifuged gradients are loaded into the station to read the RNA absorbance and protein fluorescence (optional). The resulting fractions are frozen for subsequent processing. The polysome profiles can be analyzed for changes in the area under the polysome curve.
Materials and Reagents
- Syringe, 50 ml, Luer (Thermo Fisher Scientific, catalog number: S7510-50 , Stability: Room Temperature)
- Ultracentrifuge Tube, 14 x 89 mm, Open-Top Polyclear (Seton Scientific, catalog number: 7030 , Stability: Room Temperature)
- Tube, Conical Screw Cap, 50 ml (USA Scientific, catalog number: 1500-1811 , Stability: Room Temperature)
- Tube, Microcentrifuge, 1.5 ml (USA Scientific, catalog number: 1615-5599 , Stability: Room Temperature)
- Disposable Sterile Bottle-Top Filter 0.22 µm (Corning, catalog number: 431118 )
- Caenorhabditis elegans
- cOmplete Mini, EDTA-free, Protease Inhibitor (Roche, catalog number: 11836170001 , Stability: > 1 year at 2-8 °C)
- Cycloheximide (Research Products International, catalog number: 9880 , Stability: > 2 years at -20 °C)
- EGTA (Research Products International, catalog number: E57060-25.0 , Stability: Room Temperature)
- Magnesium Chloride Hexahydrate (Thermo Fisher Scientific, catalog number: M35-500 , Stability: Room Temperature)
- Magnesium Sulfate Heptahydrate (Sigma-Aldrich, catalog number: M1880-500G , Stability: Room Temperature)
- Pestle for 1.5 ml Microcentrifuge Tubes (USA Scientific, catalog number: 1615-5599 , Stability: Room Temperature)
- Pipet tip, TipOne Ultra Low Retention Filter Tip, sterile, 200 µl (USA Scientific, catalog number: 1180-8810 , Stability: Room Temperature)
- Potassium Phosphate Monobasic (Research Products International, catalog number: P41200-500.0 , Stability: Room Temperature)
- RNase Inhibitor, SUPERase•In, 2,500 U/20 µl (Thermo Fisher Scientific, catalog number: AM2696 , Stability: Unknown)
- RNaseZap (Thermo Fisher Scientific (Invitrogen/Ambion), catalog number: 9780 , Stability: Unknown)
- Sodium Chloride (Research Products International, catalog number: S23020-5000.0 , Stability: Room Temperature)
- Sodium Deoxycholate (Sigma-Aldrich, catalog number: 30970-25G , Stability: > 2 years at -20 °C)
- Sodium Phosphate Dibasic (Sigma-Aldrich, catalog number: S3264-500G , Stability: Room Temperature)
- Sucrose (Research Products International, catalog number: S24060-1000.0 , Stability: Room Temperature)
- Tris-HCl (Research Products International, catalog number: T60050-500.0 , Stability: Room Temperature)
- Triton X-100 (Sigma-Aldrich, catalog number: T-6878 , Stability: Room Temperature)
- 0.5 M EGTA Stock Solution (see Recipes)
- 1 M MgSO4 Stock Solution (see Recipes)
- M9 Buffer (see Recipes)
- Worm Solubilization Buffer (see Recipes)
- 10x High-Salt Resolving Buffer (see Recipes)
- Sucrose Solution in High-Salt Resolving Buffer (see Recipes)
Equipment
- Balance, Digital, TS-T Series Advanced Laboratory (Thomas Scientific, catalog number: 1160T82 )
- Centrifuge, Bench top, Refrigerated, Eppendorf, 5430R (Thomas Scientific, catalog number: 1227T47 )
- Cordless Homogenizer Unit (Thomas Scientific, catalog number: 1191H98 )
- Filters, Set of LED Excitation and 2 Emission filters for EGFP fluorescence (BioComp Instruments, catalog number: FC-2-EGFP )
- Flow cell, Triax, with FC-1 monitor for dual UV and fluorescence scans (BioComp Instruments, catalog number: FC-2-UV/VIS -260 )
- Fractionator, Piston Gradient/Gradient Station (BioComp Instruments, Model 152/153 ), alternatively fractionation of samples be achieved using a Density Gradient Fractionation System (Teledyne ISCO, catalog numbers: 68-1610-010 , 60-3877-060 , 60-0084-054 , 68-1140-006 , 68-0940-016 )
- Pipettes, Set of 4, ErgoOne, Single Channel (USA Scientific, catalog number: 7104-2521 )
- 14-gauge cannula, 4” (Thomas Scientific, catalog number: 8957G54 )
- Platform Rocker, Vari-Mix (Thermo Fisher Scientific, catalog number: M79735Q )
- Rotor, TH-641 (Thermo Fisher Scientific, catalog number: NC1235764 )
- Ultracentrifuge, Sorvall, WX 80 Plus (Thermo Fisher Scientific, catalog number: NC1235763 )
- Vortexer V-32 (Thomas Scientific, catalog number: 1154J82 )
- -80 °C freezer (Thermo Scientific Revco Elite Plus, catalog number: ULT2586-6-D )
Software
- R version 3.6.1, The R Project for Statistical Computing, https://www.r-project.org/
- The provided R script 'PolysomeProfileAUC.R'
- FlowCell version 1.56, BioComp Instruments, http://biocompinstruments.com
Procedure
文章信息
版权信息
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
如何引用
Shaffer, D. and Rollins, J. A. (2020). Fluorescent Polysome Profiling in Caenorhabditis elegans . Bio-protocol 10(17): e3742. DOI: 10.21769/BioProtoc.3742.
分类
发育生物学 > 细胞生长和命运决定 > 成熟衰老
细胞生物学 > 细胞器分离 > 多聚核糖体
生物化学 > RNA > RNA-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
提问指南
+ 问题描述
写下详细的问题描述,包括所有有助于他人回答您问题的信息(例如实验过程、条件和相关图像等)。
Share
Bluesky
X
Copy link